4R0C
 
 | | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology | | Descriptor: | AbgT putative transporter family, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Su, C.-C, Bolla, J.R, Yu, E.W. | | Deposit date: | 2014-07-30 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.963 Å) | | Cite: | Crystal structure of the Alcanivorax borkumensis YdaH transporter reveals an unusual topology.
Nat Commun, 6, 2015
|
|
3V78
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3V6G
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
3T6N
 
 | |
8ORR
 
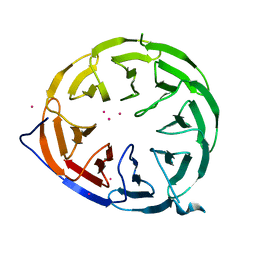 | |
7JZ2
 
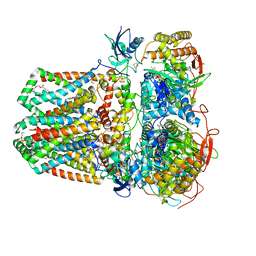 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7JZ3
 
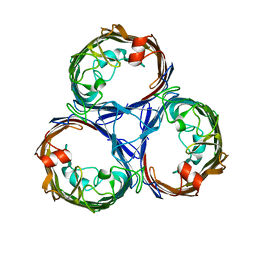 | | Osmoporin OmpC from E.coli K12 | | Descriptor: | Outer membrane protein C | | Authors: | Lyu, M, Su, C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
4R1I
 
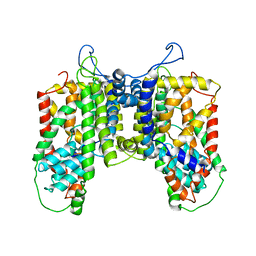 | |
4K7R
 
 | | Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation | | Descriptor: | (2S)-1-(pentanoyloxy)propan-2-yl hexanoate, Cation efflux system protein CusC | | Authors: | Su, C.-C, Lei, H.-T, Bolla, J.R, Yu, E.W. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of CusC Review Conformational Changes Accompanying Folding and Transmembrane Channel Formation.
J.Mol.Biol., 426, 2014
|
|
4MT0
 
 | |
4MT1
 
 | |
7OG0
 
 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFZ
 
 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFW
 
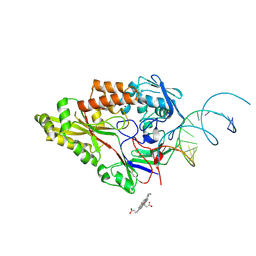 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OT9
 
 | |
8CR1
 
 | | Homo sapiens Get1/Get2 heterotetramer in complex with a Get3 dimer | | Descriptor: | ATPase ASNA1, Guided entry of tail-anchored proteins factor CAMLG,Guided entry of tail-anchored proteins factor 1,GET2-GET1, ZINC ION | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Sinning, I. | | Deposit date: | 2023-03-07 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The GET insertase exhibits conformational plasticity and induces membrane thinning.
Nat Commun, 14, 2023
|
|
5T0O
 
 | | Crystal Structure of a membrane protein | | Descriptor: | CmeB | | Authors: | Su, C.-C, Yu, E.W. | | Deposit date: | 2016-08-16 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump.
Nat Commun, 8, 2017
|
|
6SO5
 
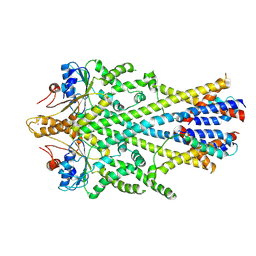 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
7JZ6
 
 | |
7JZH
 
 | |
4K7K
 
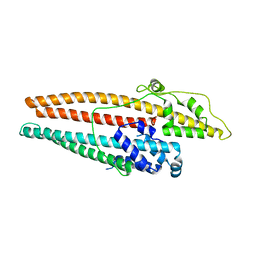 | |
4K34
 
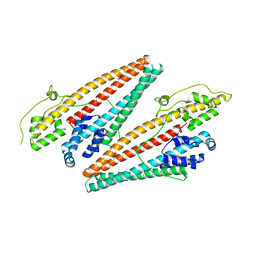 | |
6BHP
 
 | |
6WTI
 
 | | The Cryo-EM structure of the ubiquinol oxidase from Escherichia coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome o ubiquinol oxidase, ... | | Authors: | Su, C.-C. | | Deposit date: | 2020-05-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
6WU0
 
 | |
