7PUR
 
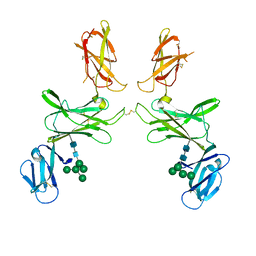 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P21 crystal form 2 | | Descriptor: | Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2021-09-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
7R3N
 
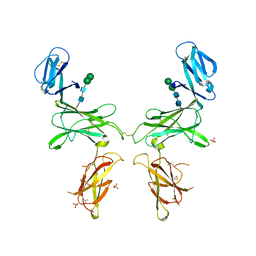 | |
5MXA
 
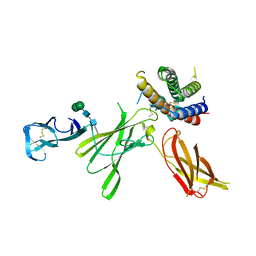 | | Structure of unbound Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-01-22 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
7PNN
 
 | |
6SMC
 
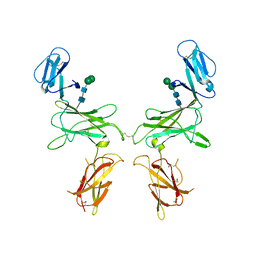 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P1 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
6SP3
 
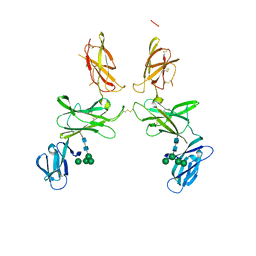 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group P21 crystal form 1 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Around she goes: the structure of mouse Interleukin-12 p80
To Be Published
|
|
5MZV
 
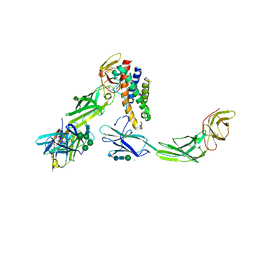 | | IL-23:IL-23R:Nb22E11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-02-01 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
5N2K
 
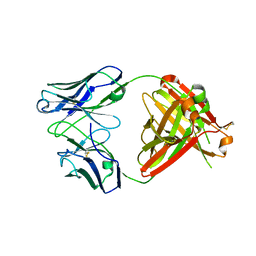 | | Structure of unbound Briakinumab FAb | | Descriptor: | 1,2-ETHANEDIOL, Briakinumab FAb heavy chain, Briakinumab FAb light chain | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-02-07 | | Release date: | 2018-01-03 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.219 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
5NJD
 
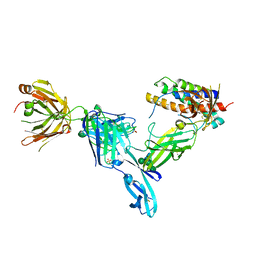 | | Structure of Interleukin 23 in complex with Briakinumab FAb | | Descriptor: | Briakinumab FAb Heavy chain, Briakinumab FAb Light chain, Interleukin-12 subunit beta, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-03-28 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
8C7L
 
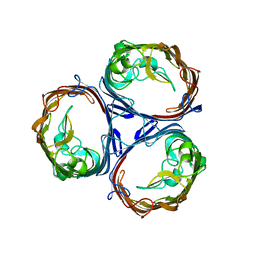 | |
8C7E
 
 | |
8C6Z
 
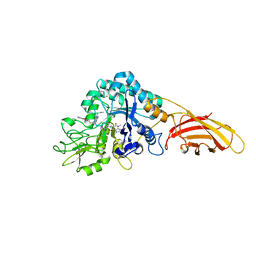 | | necrotic enteritis associated Clostridium p. chitinase F8UNI4 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), CADMIUM ION, COBALT (II) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Contribution and structural characterization of Chitinases to Clostridium perfringens Pathogenesis in Broilers
To Be Published
|
|
8C7M
 
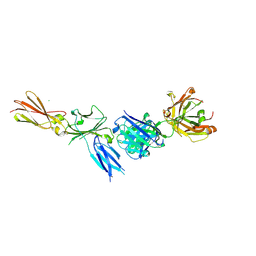 | |
8CR8
 
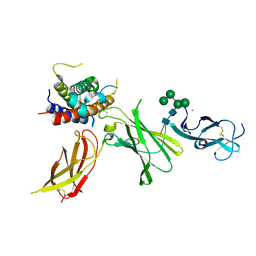 | | human Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, TERBIUM(III) ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-08 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 2024
|
|
8OSE
 
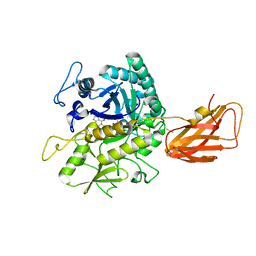 | | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C | | Descriptor: | 1,1'-PROPANE-1,3-DIYLBIS(3,7-DIMETHYL-3,7-DIHYDRO-1H-PURINE-2,6-DIONE), 1,2-ETHANEDIOL, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-18 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | C. perfringens chitinase CP4_3455 in complex with inhibitor bisdionin C
To Be Published
|
|
8OTB
 
 | |
8ODX
 
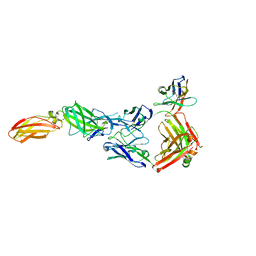 | |
8OE4
 
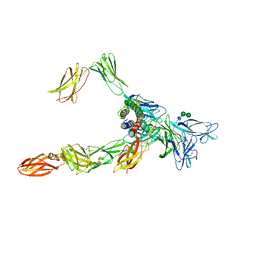 | | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 2024
|
|
8OWF
 
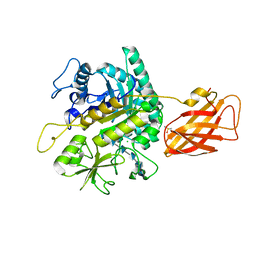 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Clostridium perfringens chitinase CP4_3455 with chitosan
To Be Published
|
|
8OYE
 
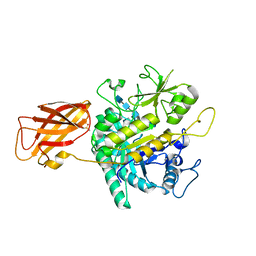 | | Clostridium perfringens chitinase CP4_3455 E196Q with chitin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitodextrinase, DIMETHYL SULFOXIDE | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-05-04 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clostridium perfringens chitinase CP4_3455 E196Q with chitin
To Be Published
|
|
8OVR
 
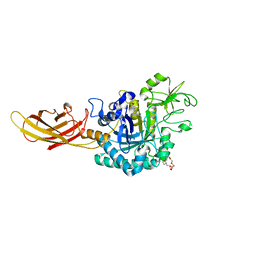 | |
8PPM
 
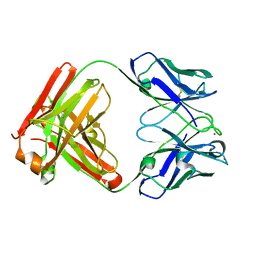 | |
8QMG
 
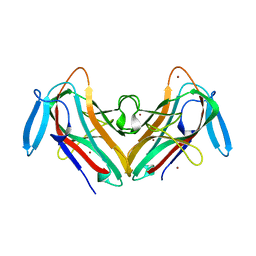 | |
8AD2
 
 | | Tobacco lectin Nictaba in complex with triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Nictaba, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tobacco lectin Nictaba in complex with triacetylchitotriose (CASP target)
To Be Published
|
|
6SFF
 
 | | mouse Interleukin-12 subunit beta - p80 homodimer in space group I41 | | Descriptor: | CHLORIDE ION, Interleukin-12 subunit beta, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homogeneously N-glycosylated proteins derived from the GlycoDelete HEK293 cell line enable diffraction-quality crystallogenesis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
