9MGY
 
 | | Cryo-EM structure of Human NLRP3 complex with compound 1 | | Descriptor: | (2M)-2-(6-{[(3R)-1-methylpiperidin-3-yl]amino}pyridazin-3-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X, Prieri, M.L.C, Bontempi, L, Embrechts, S, Paesmans, I, Bassi, M, Bhattacharya, A, Roman, S.C, Hoog, S.D, Demin, S, Gijsen, H.J.M, Hache, G, Jacobs, T, Jerhaoui, S, Leenaerts, J, Lutter, F.H, Matico, R, Oehlrich, D, Perrier, M, Ryabchuk, P, Schepens, W, Sharma, S, Somers, M, Suarez, J, Surkyn, M, Opdenbosch, N.V, Verhulst, T, Bottelbergs, A. | | Deposit date: | 2024-12-11 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
9MIG
 
 | | Cryo-EM structure of Human NLRP3 complex with compound 3 | | Descriptor: | (2P)-2-(4-{[(3R)-1-methylpiperidin-3-yl]amino}-5,6,7,8-tetrahydrophthalazin-1-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X, Prieri, M.L.C, Bontempi, L, Embrechts, S, Paesmans, I, Bassi, M, Bhattacharya, A, Roman, S.C, Hoog, S.D, Demin, S, Gijsen, H.J.M, Hache, G, Jacobs, T, Jerhaoui, S, Leenaerts, J, Lutter, F.H, Matico, R, Oehlrich, D, Perrier, M, Ryabchuk, P, Schepens, W, Sharma, S, Somers, M, Suarez, J, Surkyn, M, Opdenbosch, N.V, Verhulst, T, Bottelbergs, A. | | Deposit date: | 2024-12-12 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
4KUJ
 
 | | Structural and functional characterization of a novel Alpha Kinase from Entamoeba histolytica | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protein kinase, putative, ... | | Authors: | Rehman, S.A.A, Tarique, K.F, Bhattacharya, A, Gourinath, S, Mansuri, M.S. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional characterization of a novel Alpha Kinase from Entamoeba histolytica
To be Published
|
|
5XYL
 
 | |
4NL0
 
 | | Structural and functional characterization of a novel Alpha Kinase in complex with ADP from Entamoeba histolytica | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein kinase, putative, ... | | Authors: | Tarique, K.F, Rehman, S.A.A, Bhattacharya, A, Gourinath, S, Mansuri, M.S. | | Deposit date: | 2013-11-13 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural and functional characterization of a novel Alpha Kinase in complex with ADP from Entamoeba histolytica
To be Published
|
|
6U6X
 
 | | Human SAMHD1 bound to deoxyribo(C*G*C*C*T)-oligonucleotide | | Descriptor: | DNA SC-GS-SC-SC-DT, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6U6Y
 
 | | Human SAMHD1 bound to ribo(CGCCU)-oligonucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, RNA CGCCU, ... | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
1JFK
 
 | | MINIMUM ENERGY REPRESENTATIVE STRUCTURE OF A CALCIUM BOUND EF-HAND PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
1JFJ
 
 | | NMR SOLUTION STRUCTURE OF AN EF-HAND CALCIUM BINDING PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
2M7K
 
 | |
2LC5
 
 | | Calmodulin-like Protein from Entamoeba histolytica: Solution Structure and Calcium-Binding Properties of a Partially Folded Protein | | Descriptor: | CALCIUM ION, Calmodulin, putative | | Authors: | Rout, A.K, Padhan, N, Barnwal, R.P, Bhattacharya, A, Chary, K.V. | | Deposit date: | 2011-04-23 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Calmodulin like protein from Entamoeba histolytica: solution structure and calcium binding properties of a partially folded protein.
Biochemistry, 50, 2010
|
|
2M7M
 
 | | N-terminal domain of EhCaBP1 structure | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
2M7N
 
 | | C-terminal structure of (Y81F)-EhCaBP1 | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
4QNB
 
 | |
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6YVA
 
 | | PLpro-C111S with mISG15 | | Descriptor: | Replicase polyprotein 1a, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity.
Nature, 587, 2020
|
|
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
9MIE
 
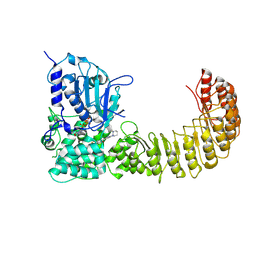 | | Human NLRP3 complex with compound 2 in the closed hexamer | | Descriptor: | (2P)-2-(4-{[(3R)-1-methylpiperidin-3-yl]amino}-6,7-dihydro-5H-cyclopenta[d]pyridazin-1-yl)-5-(trifluoromethyl)phenol, ADENOSINE-5'-TRIPHOSPHATE, NACHT, ... | | Authors: | Mammoliti, O, Carbajo, R.J, Perez-Benito, L, Yu, X. | | Deposit date: | 2024-12-12 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Discovery of Potent and Brain-Penetrant Bicyclic NLRP3 Inhibitors with Peripheral and Central In Vivo Activity.
J.Med.Chem., 68, 2025
|
|
9FGM
 
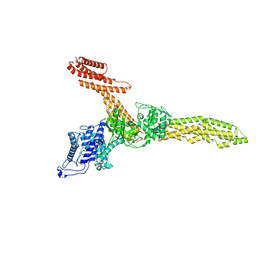 | | Cryo-EM structure of Legionella effector SdeC (3D flexible refinement) | | Descriptor: | SdeC | | Authors: | Weng, T.-H, Misra, M, Chen, W, Safarian, S, Kudryashev, M, Dikic, I. | | Deposit date: | 2024-05-24 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | BAR-like C-terminal domain of Legionella SidE family is crucial for its membrane localization and secretion
To Be Published
|
|
9FGJ
 
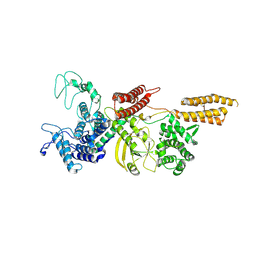 | | Cryo-EM structure of Legionella effector SdeC (PDE-mART domain) | | Descriptor: | SdeC | | Authors: | Weng, T.-H, Misra, M, Chen, W, Safarian, S, Kudryashev, M, Dikic, I. | | Deposit date: | 2024-05-24 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | BAR-like C-terminal domain of Legionella SidE family is crucial for its membrane localization and secretion
To Be Published
|
|
4LKA
 
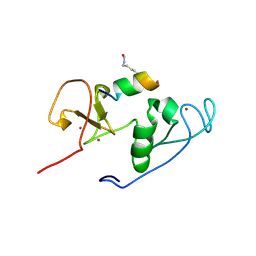 | | Crystal Structure of MOZ double PHD finger histone H3K9ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LLB
 
 | | Crystal Structure of MOZ double PHD finger histone H3K14ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LJN
 
 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
