3J7H
 
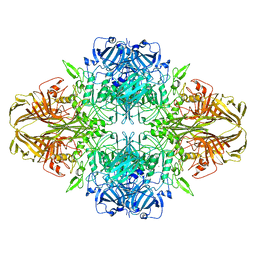 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5A1A
 
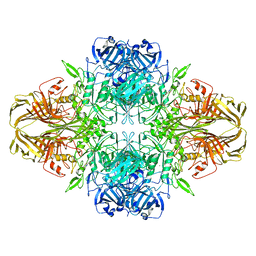 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
4CC8
 
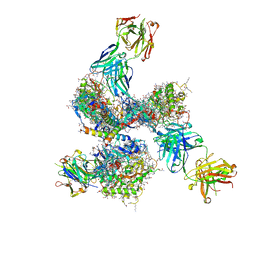 | | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | | Descriptor: | GP120, GP41, MONOCLONAL ANTIBODY VRC03 FAB HEAVY CHAIN, ... | | Authors: | Bartesaghi, A, Merk, A, Borgnia, M.J, Milne, J.L.S, Subramaniam, S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2YNJ
 
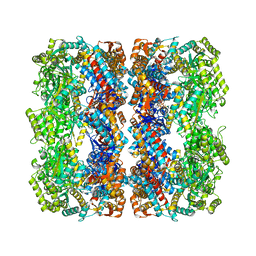 | | GroEL at sub-nanometer resolution by Constrained Single Particle Tomography | | Descriptor: | 60 KDA CHAPERONIN, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Bartesaghi, A, Lecumberry, F, Sapiro, G, Subramaniam, S. | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Structure, 20, 2012
|
|
6CVM
 
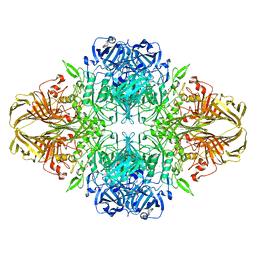 | | Atomic resolution cryo-EM structure of beta-galactosidase | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
3JD2
 
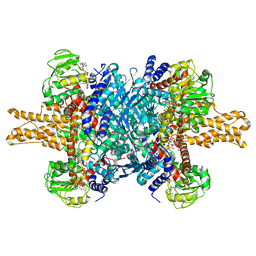 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
5K0Z
 
 | | Cryo-EM structure of lactate dehydrogenase (LDH) in inhibitor-bound state | | Descriptor: | L-lactate dehydrogenase B chain | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K12
 
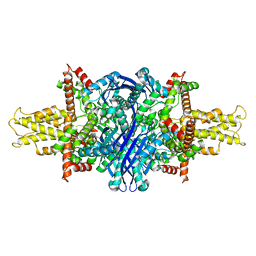 | | Cryo-EM structure of glutamate dehydrogenase at 1.8 A resolution | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K10
 
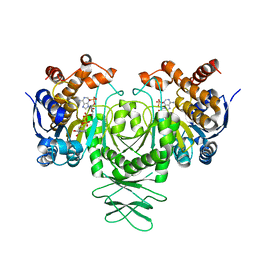 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K11
 
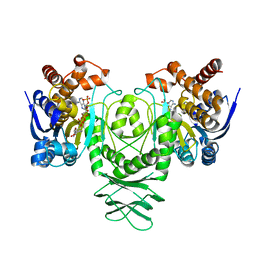 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
6EBK
 
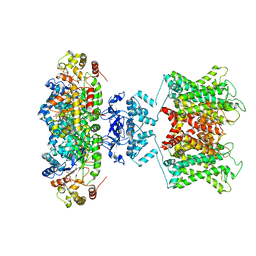 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EBL
 
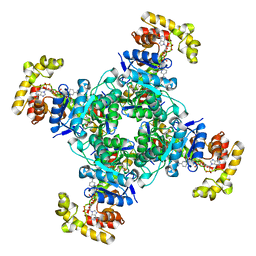 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
4UQJ
 
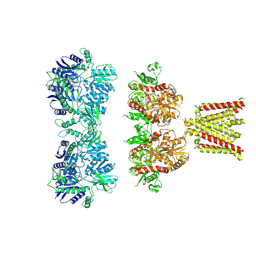 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQK
 
 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQ6
 
 | | Electron density map of GluA2em in complex with LY451646 and glutamate | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQQ
 
 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
3DNL
 
 | | Molecular structure for the HIV-1 gp120 trimer in the b12-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DNO
 
 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DNN
 
 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
5FTM
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | Authors: | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
