2KWT
 
 | | Solution structure of NS2 [27-59] | | Descriptor: | Protease NS2-3 | | Authors: | Montserret, R, Bartenschlager, R, Penin, F. | | Deposit date: | 2010-04-19 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly.
Plos Pathog., 6, 2010
|
|
2KWZ
 
 | | Solution structure of NS2 [60-99] | | Descriptor: | Protease NS2-3 | | Authors: | Montserret, R, Bartenschlager, R, Penin, F. | | Deposit date: | 2010-04-22 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly.
Plos Pathog., 6, 2010
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
5IDK
 
 | |
2LVG
 
 | |
8C0O
 
 | |
6HT4
 
 | |
8AAC
 
 | |
2XXD
 
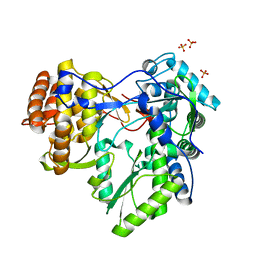 | | HCV-JFH1 NS5B polymerase structure at 1.9 angstrom | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2XYM
 
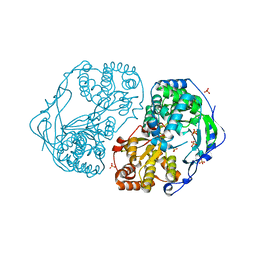 | | HCV-JFH1 NS5B T385A mutant | | Descriptor: | PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Simister, P.C, Caillet-Saguy, C, Bressanelli, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2XWH
 
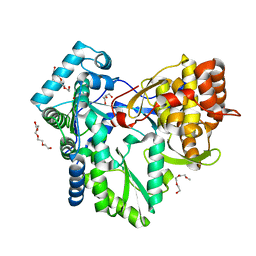 | | HCV-J6 NS5B polymerase structure at 1.8 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, POLYETHYLENE GLYCOL (N=34), RNA DEPENDENT RNA POLYMERASE | | Authors: | Scrima, N, Bressanelli, S. | | Deposit date: | 2010-11-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
2JY0
 
 | |
6YD7
 
 | |
6YD4
 
 | |
6YD3
 
 | |
6YD2
 
 | |
6ZWV
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-28 | | Release date: | 2020-08-05 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
