1H8B
 
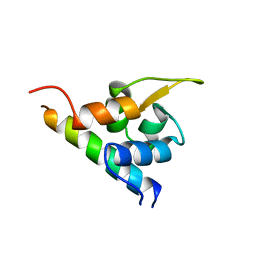 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
1DW5
 
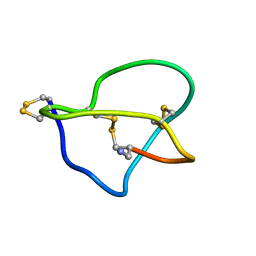 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: NO CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1DW4
 
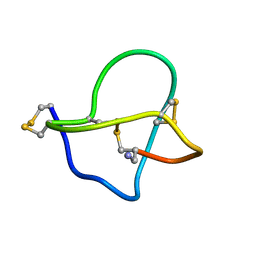 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: CONSTRAINTS ON DISULPHIDE BRIDGES | | Descriptor: | OMEGA-CONOTOXIN MVIIA | | Authors: | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | Deposit date: | 2000-01-24 | | Release date: | 2000-03-01 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
2ECH
 
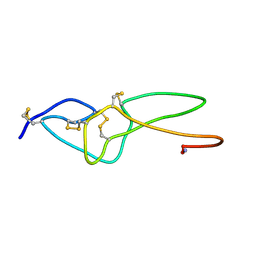 | |
2KPK
 
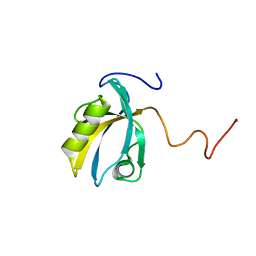 | | MAGI-1 PDZ1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
2KPL
 
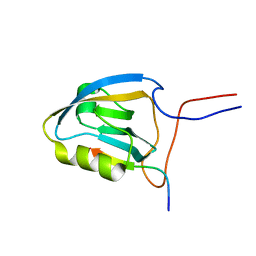 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
6I9B
 
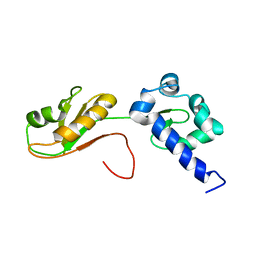 | | NMR structure of the La module from human LARP4A | | Descriptor: | La-related protein 4 | | Authors: | Conte, M.R, Martino, L, Atkinson, R.A, Kelly, G, Cruz-Gallardo, I, De Tito, S, Trotta, R. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA.
Nucleic Acids Res., 47, 2019
|
|
2MA9
 
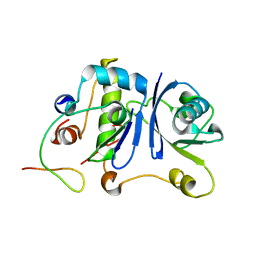 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
1RGW
 
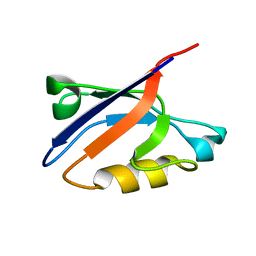 | | Solution Structure of ZASP's PDZ domain | | Descriptor: | ZASP protein | | Authors: | Au, Y, Atkinson, R.A, Pallavicini, A, Joseph, C, Martin, S.R, Muskett, F.W, Guerrini, R, Faulkner, G, Pastore, A. | | Deposit date: | 2003-11-13 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ZASP PDZ Domain; Implications for Sarcomere Ultrastructure and Enigma Family Redundancy.
Structure, 12, 2004
|
|
3ZTG
 
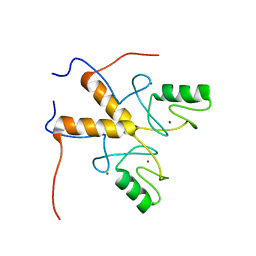 | | Solution structure of the RING finger-like domain of Retinoblastoma Binding Protein-6 (RBBP6) | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE RBBP6, ZINC ION | | Authors: | Kappo, M.A, Ab, E, Atkinson, R.A, Faro, A, Muleya, V, Mulaudzi, T, Poole, J.O, McKenzie, J.M, Pugh, D.J.R. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ring Finger-Like Domain of Retinoblastoma Binding Protein-6 (Rbbp6) Suggests It Functions as a U-Box
J.Biol.Chem., 287, 2012
|
|
2JNJ
 
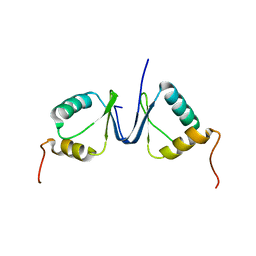 | | Solution structure of the p8 TFIIH subunit | | Descriptor: | TFIIH basal transcription factor complex TTD-A subunit | | Authors: | Vitorino, M, Atkinson, R.A, Moras, D, Poterszman, A, Kieffer, B, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-01-26 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Self-association Properties of the p8 TFIIH Subunit Responsible for Trichothiodystrophy
J.Mol.Biol., 368, 2007
|
|
5N7Y
 
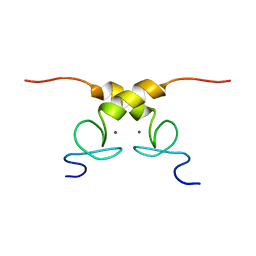 | | Solution structure of B. subtilis Sigma G inhibitor CsfB | | Descriptor: | Anti-sigma-G factor Gin, ZINC ION | | Authors: | Martinez-Lumbreras, S, Alfano, C, Atkinson, A, Isaacson, R.L. | | Deposit date: | 2017-02-21 | | Release date: | 2018-02-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Insights into Bacillus subtilis Sigma Factor Inhibitor, CsfB.
Structure, 26, 2018
|
|
6I5G
 
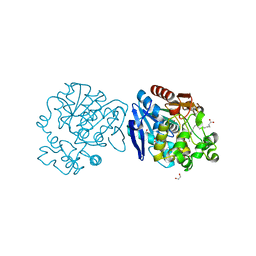 | | X-ray structure of human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD)in complex with 15d-PGJ2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
6I5E
 
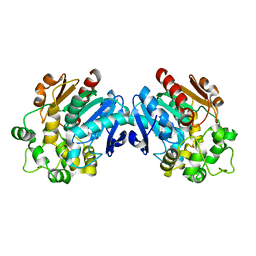 | | X-ray structure of apo human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD) | | Descriptor: | Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
4LOO
 
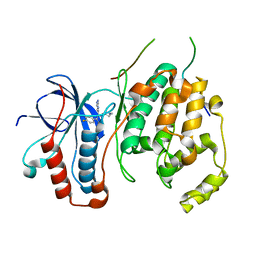 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Monoclinic crystal form) | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LOQ
 
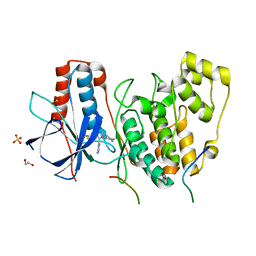 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form with bound sulphate) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, Mitogen-activated protein kinase 14, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Yue, W.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4LOP
 
 | | Structural basis of autoactivation of p38 alpha induced by TAB1 (Tetragonal crystal form) | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, L(+)-TARTARIC ACID, ... | | Authors: | Chaikuad, A, DeNicola, G.F, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Marber, M.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanism and consequence of the autoactivation of p38 alpha mitogen-activated protein kinase promoted by TAB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MTG
 
 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2FK4
 
 | |
6GIK
 
 | | NMR structure of temporin B L1FK in SDS micelles | | Descriptor: | temporinB_L1FK | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GS5
 
 | | NMR structure of temporin L in SDS micelles | | Descriptor: | Temporin-L | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GS9
 
 | | NMR structure of aurein 2.5 in SDS micelles | | Descriptor: | Aurein 2.5 | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GIJ
 
 | | NMR structure of temporin B KKG6A in SDS micelles | | Descriptor: | temporinB_KKG6A | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
