6SXI
 
 | | Antibody-anti-idiotype complex: AP33 Fab (hepatitis C virus E2 antibody) - B2.1A scFv (anti-idiotype) | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Taylor, G.L, Potter, J.A, Fadda, V, Patel, A.H, Owsianka, A.M, Cowtan, V.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-10-07 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of a structural epitope mimic: an idiotypic approach to HCV vaccine design.
NPJ Vaccines, 6, 2021
|
|
2SIM
 
 | | THE STRUCTURES OF SALMONELLA TYPHIMURIUM LT2 NEURAMINIDASE AND ITS COMPLEX WITH A TRANSITION STATE ANALOGUE AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SIALIDASE | | Authors: | Taylor, G.L, Crennell, S.J, Garman, E.F, Vimr, E.R, Laver, W.G. | | Deposit date: | 1994-07-15 | | Release date: | 1994-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
2SIL
 
 | | THE STRUCTURES OF SALMONELLA TYPHIMURIUM LT2 NEURAMINIDASE AND ITS COMPLEX WITH A TRANSITION STATE ANALOGUE AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | SIALIDASE | | Authors: | Taylor, G.L, Crennell, S.J, Garman, E.F, Vimr, E.R, Laver, W.G. | | Deposit date: | 1994-07-13 | | Release date: | 1994-08-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
4UXD
 
 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
4X47
 
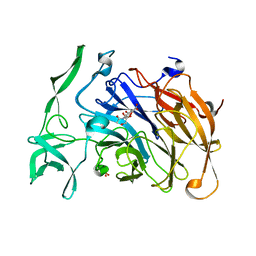 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Anhydrosialidase, PHOSPHATE ION | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
1AJ8
 
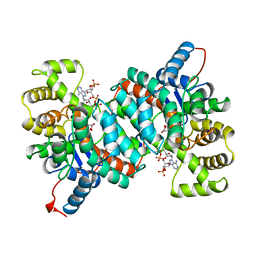 | | CITRATE SYNTHASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Ferguson, J.M.C, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
Biochemistry, 36, 1997
|
|
1A59
 
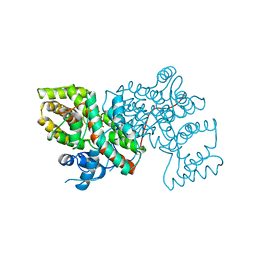 | | COLD-ACTIVE CITRATE SYNTHASE | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Gerike, U, Danson, M.J, Hough, D.W, Taylor, G.L. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium.
Structure, 6, 1998
|
|
6ER3
 
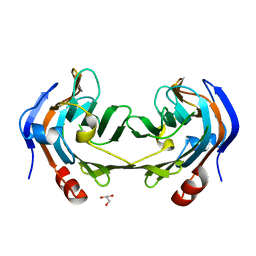 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,3 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ER4
 
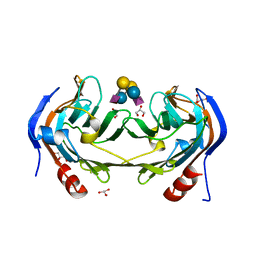 | | Ruminococcus gnavus IT-sialidase CBM40 bound to alpha2,6 sialyllactose | | Descriptor: | BNR/Asp-box repeat protein, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2017-10-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.
Nat Commun, 8, 2017
|
|
6ER2
 
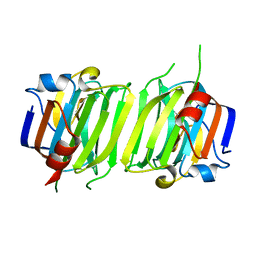 | |
4C1W
 
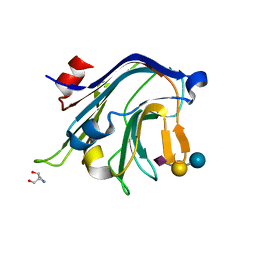 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 3'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, NEURAMINIDASE | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Prevention of Influenza by Targeting Host Receptors Using Engineered Proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1DIM
 
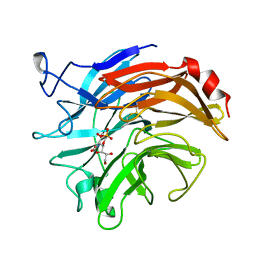 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM COMPLEXED WITH EPANA INHIBITOR | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, POTASSIUM ION, SIALIDASE | | Authors: | Garman, E.F, Crennell, S.C, Vimr, E.R, Laver, W.G, Taylor, G.L. | | Deposit date: | 1996-04-23 | | Release date: | 1996-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
1DIL
 
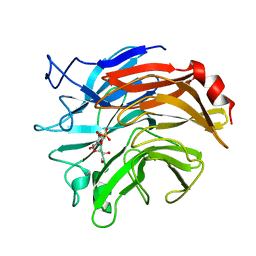 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM COMPLEXED WITH APANA AND EPANA INHIBITORS | | Descriptor: | (1R)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, (1S)-4-acetamido-1,5-anhydro-2,4-dideoxy-1-phosphono-D-glycero-D-galacto-octitol, POTASSIUM ION, ... | | Authors: | Garman, E.F, Crennell, S.C, Vimr, E.R, Laver, W.G, Taylor, G.L. | | Deposit date: | 1996-04-23 | | Release date: | 1996-12-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution.
J.Mol.Biol., 259, 1996
|
|
5BPV
 
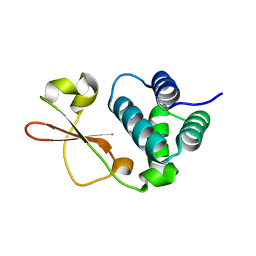 | | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Fadda, V, Cannas, V, Zinzula, L, Distinto, S, Daino, G.L, Bianco, G, Corona, A, Esposito, F, Alcaro, S, Maccioni, E, Tramontano, E, Taylor, G.L. | | Deposit date: | 2015-05-28 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure of Zaire ebolavirus VP35 RNA binding domain mutant I278A
to be published
|
|
1H0X
 
 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
4C1X
 
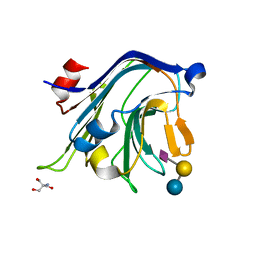 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 6'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, SIALIDASE A | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Characterization of the Carbohydrate-Binding Module of Nana Sialidase, a Pneumococcal Virulence Factor.
Bmc Struct.Biol., 15, 2015
|
|
2BF6
 
 | |
2BKY
 
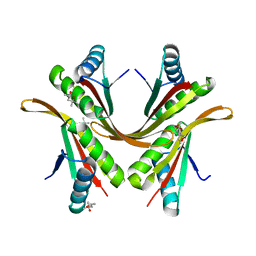 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | Authors: | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
2BER
 
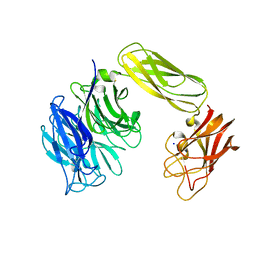 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
2CD9
 
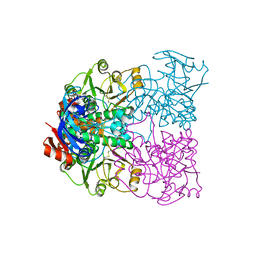 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 - apo form | | Descriptor: | GLUCOSE DEHYDROGENASE, ZINC ION | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2CDB
 
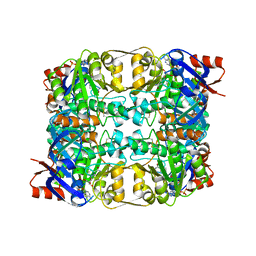 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and glucose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE 1-DEHYDROGENASE (DHG-1), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2CDC
 
 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and Xylose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE DEHYDROGENASE GLUCOSE 1-DEHYDROGENASE, DHG-1, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J. Biol. Chem., 281, 2006
|
|
2CKJ
 
 | | Human milk xanthine oxidoreductase | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pearson, A.R, Godber, B.L.J, Eisenthal, R, Taylor, G.L, Harrison, R. | | Deposit date: | 2006-04-19 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Human Milk Xanthine Dehydrogenase is Incompletely Converted to the Oxidase Form in the Absence of Proteolysis. A Structural Explanation.
To be Published
|
|
2IX2
 
 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2CDA
 
 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP | | Descriptor: | GLUCOSE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
