4YE0
 
 | | Stress-induced protein 1 truncation mutant (43 - 140) from Caenorhabditis elegans | | Descriptor: | SULFATE ION, Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
4YDZ
 
 | | Stress-induced protein 1 from Caenorhabditis elegans | | Descriptor: | Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
2Q6P
 
 | | The Chemical Control of Protein Folding: Engineering a Superfolder Green Fluorescent Protein | | Descriptor: | Green fluorescent protein mutant 3 | | Authors: | Steiner, T, Hess, P, Bae, J.H, Wiltschi, B, Moroder, L, Budisa, N. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthetic biology of proteins: tuning GFPs folding and stability with fluoroproline.
Plos One, 3, 2008
|
|
1M1G
 
 | | Crystal Structure of Aquifex aeolicus N-utilization substance G (NusG), Space Group P2(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Steiner, T, Kaiser, J.T, Marinkovic, S, Huber, R, Wahl, M.C. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of transcription factor NusG in light of its nucleic
acid- and protein-binding activities
Embo J., 21, 2002
|
|
1M1H
 
 | | Crystal structure of Aquifex aeolicus N-utilization substance G (NusG), Space group I222 | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Steiner, T, Kaiser, J.T, Marinkovic, S, Huber, R, Wahl, M.C. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities
Embo J., 21, 2002
|
|
2OEG
 
 | |
2OEF
 
 | |
1QUR
 
 | |
5K0H
 
 | |
3UTU
 
 | | High affinity inhibitor of human thrombin | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2S)-2-[(3-chloro-4-methoxybenzene)sulfonamido]-3-{[(4-cyanophenyl)methyl]carbamoyl}propanoyl]pyrrolidine-2-carboxamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Steinmetzer, T, Heine, A, Klebe, G. | | Deposit date: | 2011-11-26 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Beyond heparinization: design of highly potent thrombin inhibitors suitable for surface coupling
Chemmedchem, 7, 2012
|
|
7MLK
 
 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
7OC2
 
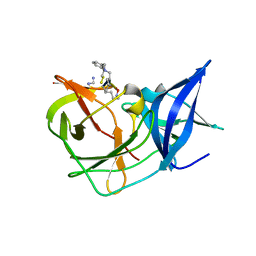 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2295 | | Descriptor: | Cyclic 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE-(7-3)-7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE-(7-19)-N-ACETYL-L-CYSTEINE-(8-25)-[3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID-()-(6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-04-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7O55
 
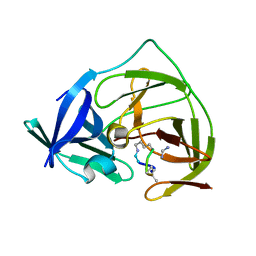 | |
7O2M
 
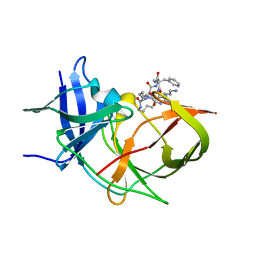 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2289 | | Descriptor: | 1-[(3~{S},6~{S},9~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-9-(phenylmethyl)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, Genome polyprotein | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7OBV
 
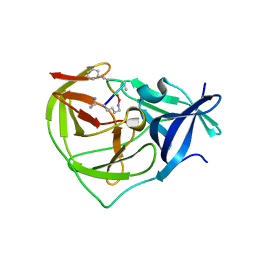 | |
7OX8
 
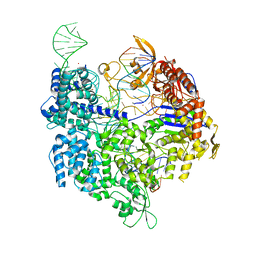 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
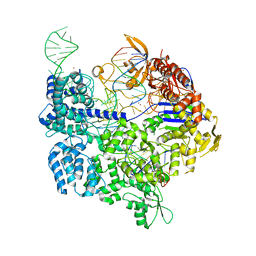 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
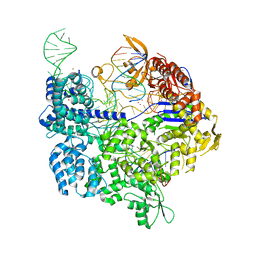 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
5EG4
 
 | | BOVINE TRYPSIN IN COMPLEX WITH CYCLIC INHIBITOR | | Descriptor: | (13S,16R)-N-(4-carbamimidoylbenzyl)-16-((N-cyclohexylsulfamoyl)amino)-3,9,15-trioxo-2,10,14-triaza-6(1,4)-piperazina-1, 11(1,4)-dibenzenacycloheptadecaphane-13-carboxamide, ACETATE ION, ... | | Authors: | Knoerlein, A, Wagner, S, Heine, A, Steinmetzer, T, Klebe, G. | | Deposit date: | 2015-10-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Optimization of Cyclic Plasmin Inhibitors: From Benzamidines to Benzylamines.
J.Med.Chem., 59, 2016
|
|
7PFY
 
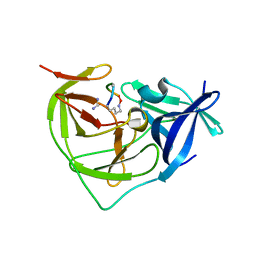 | |
7PG1
 
 | |
7PFQ
 
 | |
7PGC
 
 | |
7PFZ
 
 | |
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
