1CTJ
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sheldrick, G.M. | | Deposit date: | 1995-08-08 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ab initio determination of the crystal structure of cytochrome c6 and comparison with plastocyanin.
Structure, 3, 1995
|
|
1SHO
 
 | |
1B0Y
 
 | | MUTANT H42Q OF HIPIP FROM CHROMATIUM VINOSUM AT 0.93A | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (HIPIP) | | Authors: | Sheldrick, G.M. | | Deposit date: | 1998-11-15 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Ab initio solution and refinement of two high-potential iron protein structures at atomic resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AWD
 
 | | FERREDOXIN [2FE-2S] OXIDIZED FORM FROM CHLORELLA FUSCA | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Sheldrick, G.M. | | Deposit date: | 1997-10-01 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure determination at 1.4 A resolution of ferredoxin from the green alga Chlorella fusca
Structure Fold.Des., 7, 1999
|
|
2V9A
 
 | | Structure of Citrate-free Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | SENSOR KINASE CITA | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
3JU4
 
 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2J8T
 
 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1VTR
 
 | | STRUCTURE OF THE DEOXYTETRANUCLEOTIDE D-PAPTPAPT AND A SEQUENCE-DEPENDENT MODEL FOR POLY(DA-DT) | | Descriptor: | DNA (5'-D(*AP*TP*AP*T)-3') | | Authors: | Viswamitra, M.A, Shakked, Z, Jones, P.G, Sheldrick, G.M, Salisbury, S.A, Kennard, O. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of the Deoxytetranucleotide d-pApTpApT and a Sequence-Dependent Model for Poly(dA-dT)
Biopolymers, 21, 1982
|
|
3ITI
 
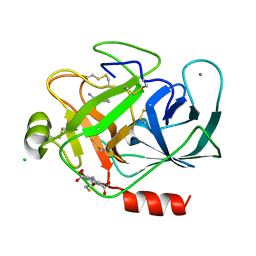 | | Structure of bovine trypsin with the MAD triangle B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Beck, T, da Cunha, C.E, Sheldrick, G.M. | | Deposit date: | 2009-08-28 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | How to get the magic triangle and the MAD triangle into your protein crystal.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6OQZ
 
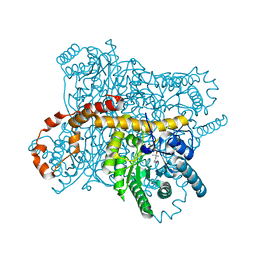 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OR0
 
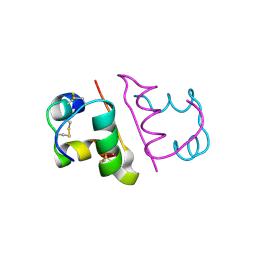 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7AF2
 
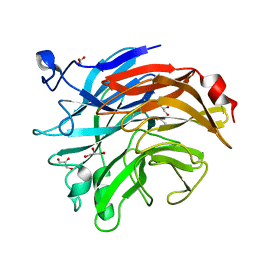 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
3SIL
 
 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Garman, E.F, Sheldrick, G.M. | | Deposit date: | 1998-07-07 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Enzyme at Atomic Resolution: The 1.05 A Structure of Salmonella Typhimurium Neuraminidase (Sialidase)
To be Published
|
|
3GT4
 
 | | Structure of proteinase K with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, SULFATE ION, proteinase K | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GT3
 
 | | Structure of proteinase K with the mad triangle B3C | | Descriptor: | 5-amino-2,4,6-tribromobenzene-1,3-dicarboxylic acid, Proteinase K, SULFATE ION | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6R99
 
 | |
2V9B
 
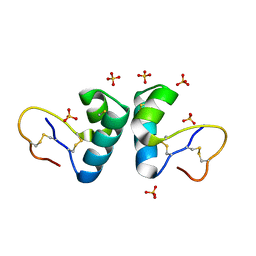 | | X-ray structure of viscotoxin B2 from Viscum album | | Descriptor: | SULFATE ION, VISCOTOXIN-B | | Authors: | Debreczeni, J.E, Pal, A, Kahle, B, Zeeck, A, Sheldrick, G.M. | | Deposit date: | 2007-08-23 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of viscotoxins A1 and B2 from European mistletoe solved using native data alone.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
2WFI
 
 | |
2WFJ
 
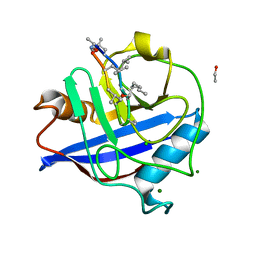 | | Atomic resolution crystal structure of the PPIase domain of human cyclophilin G in complex with cyclosporin A. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYCLOSPORIN A, ... | | Authors: | Stegmann, C.M, Sheldrick, G.M, Wahl, M.C. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | The Thermodynamic Influence of Trapped Water Molecules on a Protein-Ligand Interaction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4MIV
 
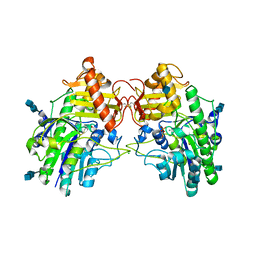 | | Crystal Structure of Sulfamidase, Crystal Form L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Sheldrick, G.M, Gaertner, J, Kraetzner, R, Steinfeld, R. | | Deposit date: | 2013-09-02 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3E3T
 
 | | Structure of porcine pancreatic elastase with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Elastase-1, IODIDE ION, ... | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E3S
 
 | | Structure of thaumatin with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3E3D
 
 | | Structure of hen egg white lysozyme with the magic triangle I3C | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Lysozyme C | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2008-08-07 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A magic triangle for experimental phasing of macromolecules
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4MHX
 
 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
