4LH6
 
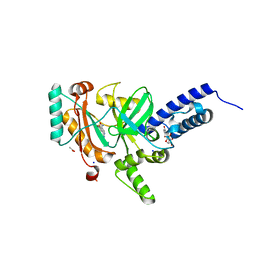 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4WF1
 
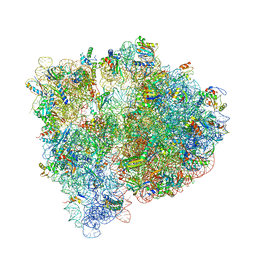 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2IQG
 
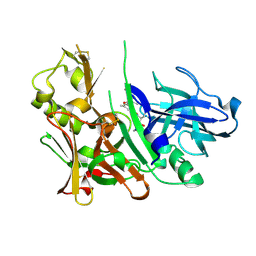 | | Crystal Structure of Hydroxyethyl Secondary Amine-based Peptidomimetic Inhibitor of Human Beta-Secretase (BACE) | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-IODOBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Benson, T.E, Woods, D.D, Prince, D.B, Tomasselli, A.G, Emmons, T.L. | | Deposit date: | 2006-10-13 | | Release date: | 2007-02-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis, and Crystal Structure of Hydroxyethyl Secondary Amine-Based Peptidomimetic Inhibitors of Human beta-Secretase.
J.Med.Chem., 50, 2007
|
|
4LH7
 
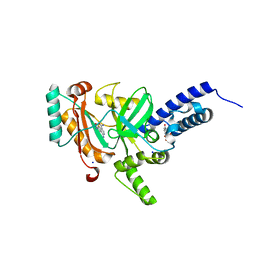 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-aminothieno[3,2-c]pyridine-2,7-dicarboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Boriack-Sjodin, P.A, Prince, D.B. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2P83
 
 | | Potent and selective isophthalamide S2 hydroxyethylamine inhibitor of BACE1 | | Descriptor: | Beta-secretase 1, N~3~-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-N~1~,N~1~-DIPROPYLBENZENE-1,3,5-TRICARBOXAMIDE, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2007-03-21 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent and selective isophthalamide S(2) hydroxyethylamine inhibitors of BACE1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4OKG
 
 | |
6B3E
 
 | | Crystal structure of human CDK12/CyclinK in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S)-1-(6-{[(4,5-difluoro-1H-benzimidazol-2-yl)methyl]amino}-9-ethyl-9H-purin-2-yl)piperidin-2-yl]ethan-1-ol, Cyclin-K, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-09-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure-Based Design of Selective Noncovalent CDK12 Inhibitors.
ChemMedChem, 13, 2018
|
|
1LRZ
 
 | | x-ray crystal structure of staphylococcus aureus femA | | Descriptor: | factor essential for expression of methicillin resistance | | Authors: | Benson, T, Prince, D, Mutchler, V, Curry, K, Ho, A, Sarver, R, Hagadorn, J, Choi, G, Garlick, R. | | Deposit date: | 2002-05-16 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of Staphylococcus aureus FemA.
Structure, 10, 2002
|
|
2HM1
 
 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor (2) | | Descriptor: | Beta-secretase 1, N-{(1S)-2-({(1S,2R)-1-(3,5-DIFLUOROBENZYL)-3-[(3-ETHYLBENZYL)AMINO]-2-HYDROXYPROPYL}AMINO)-2-OXO-1-[(PENTYLSULFONYL)METHYL]ETHYL}NICOTINAMIDE | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-07-10 | | Release date: | 2007-01-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2HIZ
 
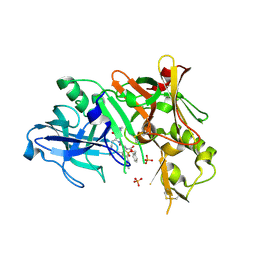 | | Crystal Structure of human beta-secretase (BACE) in the presence of an inhibitor | | Descriptor: | BENZYL [(1S)-2-({(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}AMINO)-2-OXO-1-{[(1-PROPYLBUTYL)SULFONYL]METHYL}ETHYL]CARBAMATE, Beta-secretase 1, PHOSPHATE ION | | Authors: | Benson, T.E, Prince, D.B, Tomasselli, A.G, Emmons, T.L, Paddock, D.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-01-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of potent inhibitors of human beta-secretase. Part 1.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5IT8
 
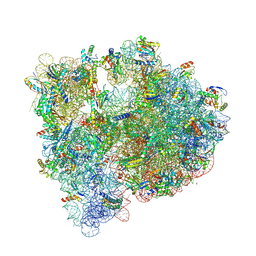 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J88
 
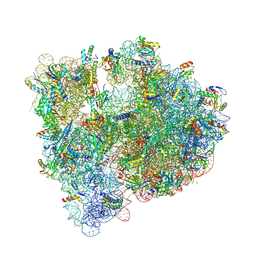 | | Structure of the E coli 70S ribosome with the U1060A mutation in 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-06 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4WZ4
 
 | | Crystal structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, GLYCEROL, {(3R)-6-[(3-amino-1,2,4-thiadiazol-5-yl)oxy]-1-hydroxy-4,5-dimethyl-1,3-dihydro-2,1-benzoxaborol-3-yl}acetic acid | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
4WZ5
 
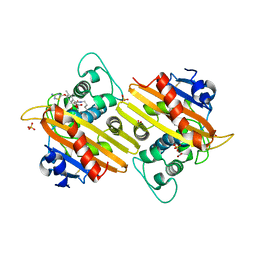 | | Crystal structure of P. aeruginosa OXA10 | | Descriptor: | Beta-lactamase OXA-10, CARBON DIOXIDE, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
Acs Infect Dis., 1, 2015
|
|
4WYY
 
 | | Crystal Structure of P. aeruginosa AmpC | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-11-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4,5-Disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles Are Broad-Spectrum Serine beta-Lactamase Inhibitors.
ACS Infect Dis, 1, 2015
|
|
3U2D
 
 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3U2K
 
 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
5N21
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | 2-[(2~{S})-1-[3-cyano-7-[(2-oxidanylidene-3,4-dihydro-1~{H}-quinolin-6-yl)amino]pyrazolo[1,5-a]pyrimidin-5-yl]pyrrolidin-2-yl]ethanoic acid, B-cell lymphoma 6 protein, CHLORIDE ION | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1Z
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | B-cell lymphoma 6 protein, CHLORIDE ION, pyrazolo-pyrimidine macrocycle | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N20
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-[(3~{R})-3-(dimethylamino)pyrrolidin-1-yl]phenyl]ethanamide | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N1V
 
 | | Crystal structure of the protein kinase CK2 catalytic subunit in complex with pyrazolo-pyrimidine macrocyclic ligand | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
4FM8
 
 | | Crystal Structure of BACE with Compound 12a | | Descriptor: | (5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-8-[3-(propan-2-yloxy)benzyl]-2-thia-1,3,8-triazaspiro[4.5]decane 2,2-dioxide, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
4FM7
 
 | | Crystal Structure of BACE with Compound 14g | | Descriptor: | 4-{[(5R,7S)-1-(3-fluorophenyl)-3,7-dimethyl-2,2-dioxido-2-thia-1,3,8-triazaspiro[4.5]dec-8-yl]methyl}-2-(propan-2-yloxy)phenol, Beta-secretase 1, ZINC ION | | Authors: | Vajdos, F.F, Varghese, A.H. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-03 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Spirocyclic sulfamides as beta-secretase 1 (BACE-1) inhibitors for the treatment of Alzheimer's disease: utilization of structure based drug design, WaterMap, and CNS penetration studies to identify centrally efficacious inhibitors.
J.Med.Chem., 55, 2012
|
|
4P73
 
 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
