1RZW
 
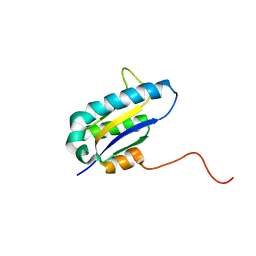 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
4AYK
 
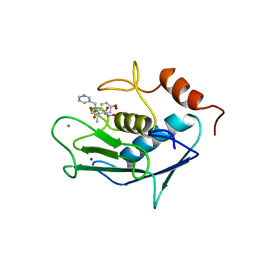 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
3AYK
 
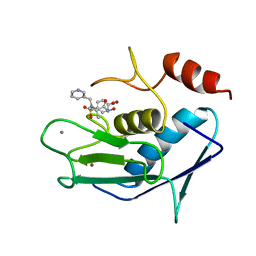 | | CATALYTIC FRAGMENT OF HUMAN FIBROBLAST COLLAGENASE COMPLEXED WITH CGS-27023A, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, N-HYDROXY-2(R)-[[(4-METHOXYPHENYL)SULFONYL](3-PICOLYL)AMINO]-3-METHYLBUTANAMIDE HYDROCHLORIDE, PROTEIN (COLLAGENASE), ... | | Authors: | Powers, R, Moy, F.J. | | Deposit date: | 1999-02-01 | | Release date: | 1999-06-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the catalytic fragment of human fibroblast collagenase complexed with a sulfonamide derivative of a hydroxamic acid compound.
Biochemistry, 38, 1999
|
|
1AYK
 
 | |
1BLA
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
1BLD
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
2AYK
 
 | |
8E2O
 
 | |
2KEW
 
 | | The solution structure of Bacillus subtilis SR211 START domain by NMR spectroscopy | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C, and (15)N NMR assignments for the Bacillus subtilis yndB START domain.
Biomol.Nmr Assign., 3, 2009
|
|
2KTE
 
 | | The solution structure of Bacillus subtilis, YndB, Northeast Structural Genomics Consoritum Target SR211 | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Montelione, G.T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of YndB, an AHSA1 protein from Bacillus subtilis.
Proteins, 78, 2010
|
|
1FLS
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
1F7X
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
1FM1
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC FRAGMENT OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED WITH A HYDROXAMIC ACID INHIBITOR | | Descriptor: | CALCIUM ION, COLLAGENASE-3, N-HYDROXY-2-[(4-METHOXY-BENZENESULFONYL)-PYRIDIN-3-YLMETHYL-AMINO]-3-METHYL-BENZAMIDE, ... | | Authors: | Moy, F.J, Chanda, P.K, Chen, J.M, Cosmi, S, Edris, W, Levin, J.I, Powers, R. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the catalytic fragment of human collagenase-3 (MMP-13) complexed with a hydroxamic acid inhibitor.
J.Mol.Biol., 302, 2000
|
|
1F7W
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
1EZT
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-11 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
1EZY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-12 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
5JN6
 
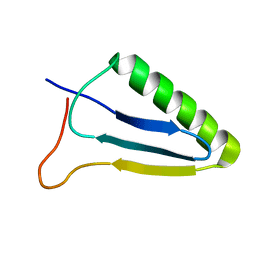 | |
1IJZ
 
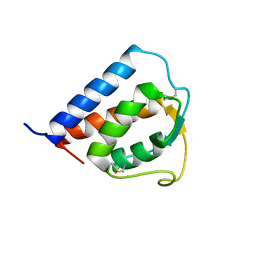 | |
1ICH
 
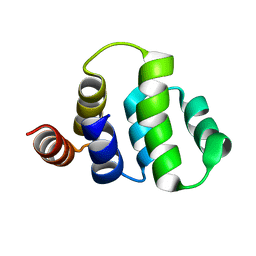 | | SOLUTION STRUCTURE OF THE TUMOR NECROSIS FACTOR RECEPTOR-1 DEATH DOMAIN | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR-1 | | Authors: | Sukits, S.F, Lin, L.-L, Malakian, K, Powers, R, Xu, G.-Y. | | Deposit date: | 2001-04-01 | | Release date: | 2002-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tumor necrosis factor receptor-1 death domain.
J.Mol.Biol., 310, 2001
|
|
1IK0
 
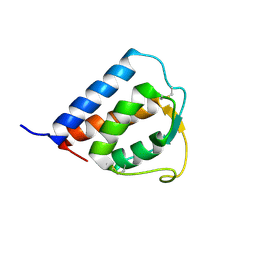 | |
2LZN
 
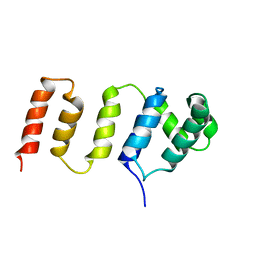 | |
1HY8
 
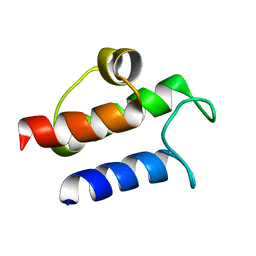 | | SOLUTION STRUCTURE OF B. SUBTILIS ACYL CARRIER PROTEIN | | Descriptor: | ACYL CARRIER PROTEIN | | Authors: | Xu, G.-Y, Tam, A, Lin, L, Hixon, J, Fritz, C.C, Power, R. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of B. subtilis acyl carrier protein.
Structure, 9, 2001
|
|
2M6Y
 
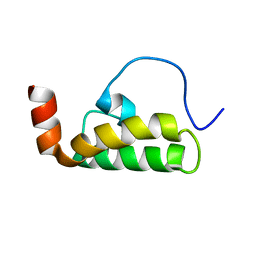 | | The solution structure of the J-domain of human DnaJA1 | | Descriptor: | DnaJ homolog subfamily A member 1 | | Authors: | Stark, J.L, Mehla, K, Chaika, N, Acton, T.B, Xiao, R, Singh, P.K, Montelione, G.T, Powers, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer.
Biochemistry, 53, 2014
|
|
2JXH
 
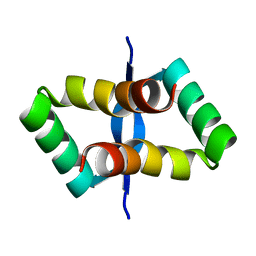 | |
2JXG
 
 | |
