6N5V
 
 | | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine | | Descriptor: | 2-(1H-indol-4-yl)ethan-1-amine, Strictosidine synthase | | Authors: | Cai, Y, Shao, N, Xie, H, Futamura, Y, Panjikar, S, Liu, H, Zhu, H, Osada, H, Zou, H. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine
to be published
|
|
8YHR
 
 | | DHODH in complex with furocoumavirin | | Descriptor: | 4-methyl-8-[(S)-oxidanyl(phenyl)methyl]-9-phenyl-furo[2,3-h]chromen-2-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Hara, K, Okumura, H, Nakahara, M, Sato, M, Hashimoto, H, Osada, H, Watanabe, K. | | Deposit date: | 2024-02-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Analyses of Inhibition of Human Dihydroorotate Dehydrogenase by Antiviral Furocoumavirin.
Biochemistry, 63, 2024
|
|
7T5J
 
 | | Crystal Structure of Strictosidine Synthase in complex with S-IPA (2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2S)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
7T5I
 
 | | Crystal Structure of Strictosidine Synthase in complex with R-IPA(2-(1H-indol-3-yl) propan-1-amine) | | Descriptor: | (2R)-2-(1H-indol-3-yl)propan-1-amine, Strictosidine synthase | | Authors: | Liu, H, Panjikar, S, Futamura, Y, Shao, N, Osada, H, Zou, H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Beta-Branched Tryptamine Provoked Non Sandwich-Like-Mode Catalysis of Strictosidine Synthase to Antimalarial Indole Alkaloids
to be published
|
|
6KOG
 
 | | Ketosynthase domain in tenuazonic acid synthetase 1 (TAS1). | | Descriptor: | GLYCEROL, Hybrid PKS-NRPS synthetase TAS1, SULFATE ION | | Authors: | Yun, C.S, Nishimoto, K, Motoyama, T, Hino, T, Nagano, S, Osada, H. | | Deposit date: | 2019-08-10 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Unique features of the ketosynthase domain in a nonribosomal peptide synthetase-polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1.
J.Biol.Chem., 295, 2020
|
|
3U0V
 
 | | Crystal Structure Analysis of human LYPLAL1 | | Descriptor: | Lysophospholipase-like protein 1 | | Authors: | Burger, M, Zimmermann, T.J, Kondoh, Y, Stege, P, Watanabe, N, Osada, H, Waldmann, H, Vetter, I.R. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases
J.Lipid Res., 53, 2012
|
|
3VNQ
 
 | |
3VNR
 
 | |
3VNS
 
 | |
5IH5
 
 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A | | Descriptor: | 6-(3-chlorophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH6
 
 | | Human Casein Kinase 1 isoform delta (kinase domain) in complex with Epiblastin A derivative | | Descriptor: | 6-(3-bromophenyl)pteridine-2,4,7-triamine, Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
5IH4
 
 | | Human Casein Kinase 1 isoform delta apo (kinase domain) | | Descriptor: | Casein kinase I isoform delta, S,R MESO-TARTARIC ACID, SULFATE ION, ... | | Authors: | Ursu, A, Illich, D.J, Takemoto, Y, Porfetye, A.T, Zhang, M, Brockmeyer, A, Janning, P, Watanabe, N, Osada, H, Vetter, I.R, Ziegler, S, Schoeler, H.R, Waldmann, H. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epiblastin A Induces Reprogramming of Epiblast Stem Cells Into Embryonic Stem Cells by Inhibition of Casein Kinase 1.
Cell Chem Biol, 23, 2016
|
|
2ZA0
 
 | | Crystal structure of mouse glyoxalase I complexed with methyl-gerfelin | | Descriptor: | Glyoxalase I, ZINC ION, methyl 4-(2,3-dihydroxy-5-methylphenoxy)-2-hydroxy-6-methylbenzoate | | Authors: | Okumura, H, Kawatani, M, Osada, H. | | Deposit date: | 2007-09-26 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZHP
 
 | | Crystal structure of bleomycin-binding protein from Streptoalloteichus hindustanus complexed with bleomycin derivative | | Descriptor: | Bleomycin resistance protein, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2008-02-06 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Affinity Relationship Study of Bleomycins and Shble protein Using a Chemical Array
To be Published
|
|
3ACL
 
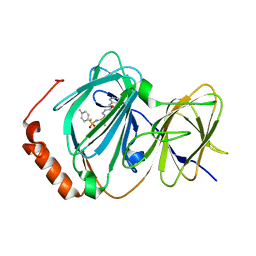 | | Crystal Structure of Human Pirin in complex with Triphenyl Compound | | Descriptor: | FE (II) ION, N-{[4-(benzyloxy)phenyl](methyl)-lambda~4~-sulfanylidene}-4-methylbenzenesulfonamide, Pirin | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2010-01-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule inhibitor shows that pirin regulates migration of melanoma cells
Nat.Chem.Biol., 6, 2010
|
|
3WVS
 
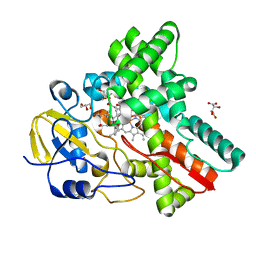 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
5INI
 
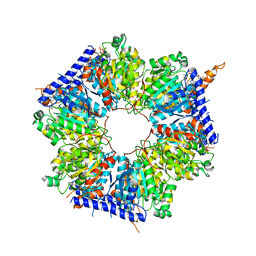 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | HEXANOYL-COENZYME A, Putative carboxyl transferase | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Milligan, J.C, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
5ING
 
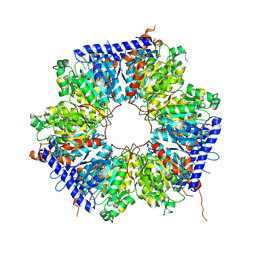 | | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender unit | | Descriptor: | Putative carboxyl transferase | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Song, L, Withall, D.M, Milligan, J.C, Takahashi, S, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
5INF
 
 | | Structural basis for acyl-CoA carboxylase-mediated assembly of unusual polyketide synthase extender units incorporated into the stambomycin antibiotics | | Descriptor: | Carboxyl transferase, HEXANOYL-COENZYME A | | Authors: | Valentic, T.R, Ray, L, Miyazawa, T, Withall, D.M, Song, L, Osada, H, Tsai, S.C, Challis, G.L. | | Deposit date: | 2016-03-07 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A crotonyl-CoA reductase-carboxylase independent pathway for assembly of unusual alkylmalonyl-CoA polyketide synthase extender units.
Nat Commun, 7, 2016
|
|
5KEJ
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
4V1D
 
 | | Ternary complex among two human derived single chain antibody fragments and Cn2 toxin from scorpion Centruroides noxius. | | Descriptor: | BETA-MAMMAL TOXIN CN2, SINGLE CHAIN ANTIBODY FRAGMENT LR, HEAVY CHAIN, ... | | Authors: | Riano-Umbarila, L, Serrano-Posada, H, Rojas-Trejo, S, Rudino-Pinera, E, Becerril, B. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Optimal Neutralization of Centruroides Noxius Venom is Understood Through a Structural Complex between Two Antibody Fragments and the Cn2 Toxin.
J.Biol.Chem., 291, 2016
|
|
6OH6
 
 | |
6OH8
 
 | |
6WNU
 
 | |
6WNI
 
 | |
