1DSB
 
 | |
5GPB
 
 | |
2GPB
 
 | |
3GPB
 
 | |
1Z1H
 
 | | HIV-1 protease complexed with macrocyclic peptidomimetic inhibitor 3 | | Descriptor: | N-{(2R)-2-HYDROXY-2-[(8S,11S)-8-ISOPROPYL-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-11-YL]ETHYL}-N-ISOPENTYLBENZENESULFONAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-04 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
4GPB
 
 | |
1HNN
 
 | | CRYSTAL STRUCTURE OF HUMAN PNMT COMPLEXED WITH SK&F 29661 AND ADOHCY(SAH) | | Descriptor: | 1,2,3,4-TETRAHYDRO-ISOQUINOLINE-7-SULFONIC ACID AMIDE, PHENYLETHANOLAMINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Martin, J.L, Begun, J, McLeish, M.J, Caine, J.M, Grunewald, G.L. | | Deposit date: | 2000-12-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT.
Structure, 9, 2001
|
|
1B6K
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 5 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2]HEPTADECA-1(16),13(17),14- TRIEN-11-YLAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL)-PROPYL]-3-METHYL-2- (2-OXO-PYRROLIDIN-1-YL)-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6P
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 7 | | Descriptor: | N-[3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YAMINO)-2-HYDROXY-1-(4-HYDROXY-BENZYL) -PROPYL]-3-METHYL-2-PROPIONYLAMINO-BUTYRAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6M
 
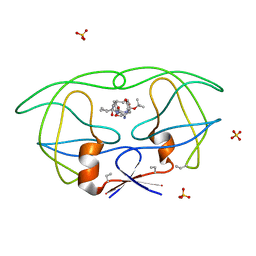 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 6 | | Descriptor: | RETROPEPSIN, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6J
 
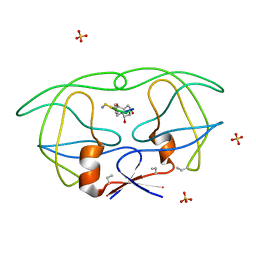 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 1 | | Descriptor: | CYCLIC PEPTIDE INHIBITOR, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
1B6L
 
 | | HIV-1 PROTEASE COMPLEXED WITH MACROCYCLIC PEPTIDOMIMETIC INHIBITOR 4 | | Descriptor: | 1-[2-(8-CARBAMOYLMETHYL-6,9-DIOXO-2-OXA-7,10-DIAZA-BICYCLO[11.2.2]HEPTADECA- 1(16),13(17),14-TRIEN-11-YL)-2-HYDROXY-ETHYL]-PIPERIDINE-2-CARBOXYLIC ACID TERT-BUTYLAMIDE, RETROPEPSIN, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 1999-01-17 | | Release date: | 2000-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease.
Biochemistry, 38, 1999
|
|
3E90
 
 | |
1A2M
 
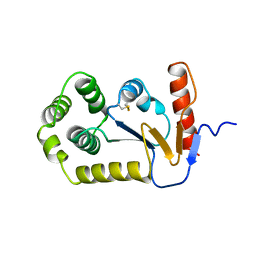 | |
1A2J
 
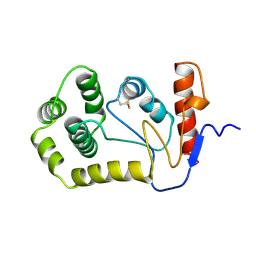 | | OXIDIZED DSBA CRYSTAL FORM II | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1A2L
 
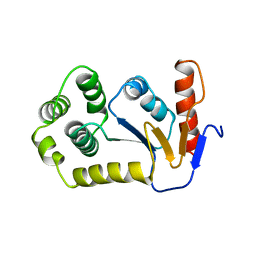 | | REDUCED DSBA AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | DISULFIDE BOND FORMATION PROTEIN | | Authors: | Martin, J.L, Guddat, L.W. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization.
Structure, 6, 1998
|
|
1FVJ
 
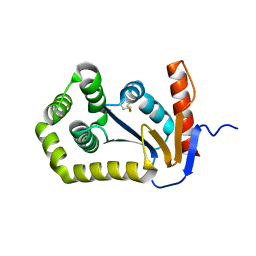 | |
1FVK
 
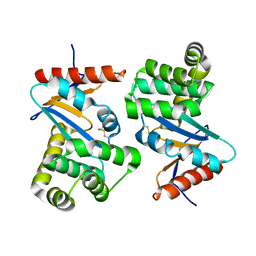 | |
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
1CPI
 
 | |
1KNG
 
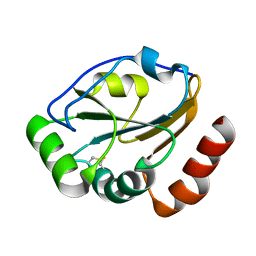 | | Crystal structure of CcmG reducing oxidoreductase at 1.14 A | | Descriptor: | THIOL:DISULFIDE INTERCHANGE PROTEIN CYCY | | Authors: | Edeling, M.A, Guddat, L.W, Fabianek, R.A, Thony-Meyer, L, Martin, J.L. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment
Structure, 10, 2002
|
|
6EEZ
 
 | |
2V1O
 
 | | Crystal structure of N-terminal domain of acyl-CoA thioesterase 7 | | Descriptor: | COENZYME A, CYTOSOLIC ACYL COENZYME A THIOESTER HYDROLASE | | Authors: | Forwood, J.K, Thakur, A.S, Guncar, G, Marfori, M, Mouradov, D, Meng, W.N, Robinson, J, Huber, T, Kellie, S, Martin, J.L, Hume, D.A, Kobe, B. | | Deposit date: | 2007-05-28 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Recruitment of Tandem Hotdog Domains in Acyl-Coa Thioesterase 7 and its Role in Inflammation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1V58
 
 | | Crystal Structure Of the Reduced Protein Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
