5IKB
 
 | | Crystal structure of the kainate receptor GluK4 ligand binding domain in complex with kainate | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Kristensen, O, Kristensen, L.B, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of a High-Affinity Kainate Receptor: GluK4 Ligand-Binding Domain Crystallized with Kainate.
Structure, 24, 2016
|
|
5DRV
 
 | |
1P9Y
 
 | |
1OMS
 
 | |
1T6C
 
 | | Structural characterization of the Ppx/GppA protein family: crystal structure of the Aquifex aeolicus family member | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kristensen, O, Laurberg, M, Liljas, A, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family
Biochemistry, 43, 2004
|
|
1T6D
 
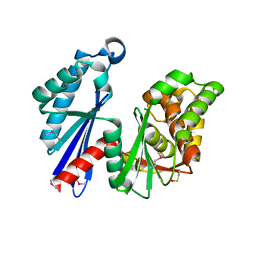 | | MIRAS phasing of the Aquifex aeolicus Ppx/GppA phosphatase: crystal structure of the type II variant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, exopolyphosphatase | | Authors: | Kristensen, O, Laurberg, M, Liljas, A, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family
Biochemistry, 43, 2004
|
|
2J4R
 
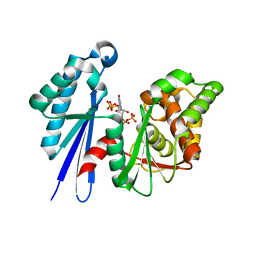 | | Structural Study of the Aquifex aeolicus PPX-GPPA enzyme | | Descriptor: | EXOPOLYPHOSPHATASE, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kristensen, O. | | Deposit date: | 2006-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the Ppx/Gppa Phosphatase from Aquifex Aeolicus in Complex with the Alarmone Ppgpp
J.Mol.Biol., 375, 2008
|
|
2FPF
 
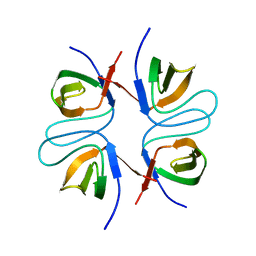 | |
2FPD
 
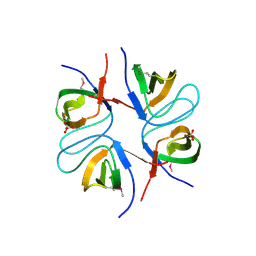 | |
1CQN
 
 | |
1CQM
 
 | |
1OB2
 
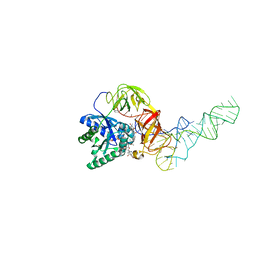 | | E. coli elongation factor EF-Tu complexed with the antibiotic kirromycin, a GTP analog, and Phe-tRNA | | Descriptor: | ELONGATION FACTOR TU, KIRROMYCIN, MAGNESIUM ION, ... | | Authors: | Kristensen, O, Nissen, P, Nyborg, J. | | Deposit date: | 2003-01-24 | | Release date: | 2004-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Kirromycin Defines a Specific Domain Arrangement of Elongation Factor EF-TU
To be Published
|
|
1QJH
 
 | |
2FPE
 
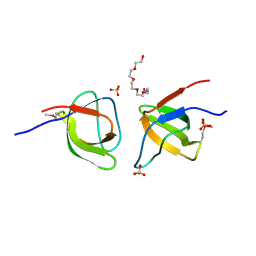 | | Conserved dimerization of the ib1 src-homology 3 domain | | Descriptor: | C-jun-amino-terminal kinase interacting protein 1, HEXAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Guenat, S, Dar, I, Bonny, C, Kastrup, J.S, Gajhede, M, Kristensen, O. | | Deposit date: | 2006-01-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique set of SH3-SH3 interactions controls IB1 homodimerization
Embo J., 25, 2006
|
|
2DQU
 
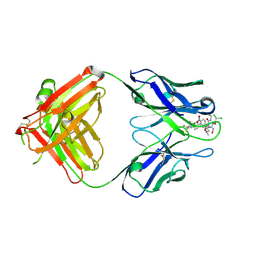 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DQT
 
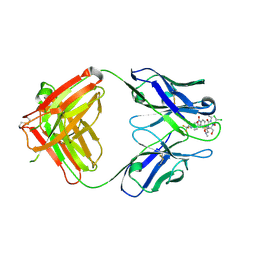 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
4ZG1
 
 | |
6FF3
 
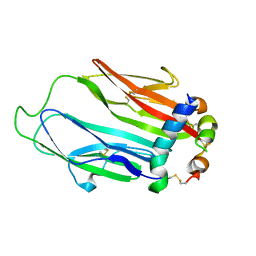 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L1 with Human IGF-I | | Descriptor: | Insulin-like growth factor I, Neural/ectodermal development factor IMP-L2 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D, Viola, C.M. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
6FEY
 
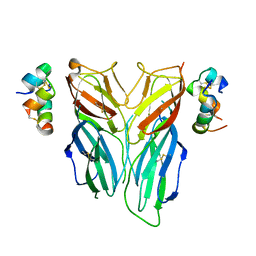 | | Crystal structure of Drosophila neural ectodermal development factor Imp-L2 with Drosophila DILP5 insulin | | Descriptor: | Neural/ectodermal development factor IMP-L2, Probable insulin-like peptide 5 | | Authors: | Brzozowski, A.M, Kulahin, N, Kristensen, O, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2018-01-03 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structures of insect Imp-L2 suggest an alternative strategy for regulating the bioavailability of insulin-like hormones.
Nat Commun, 9, 2018
|
|
4IIA
 
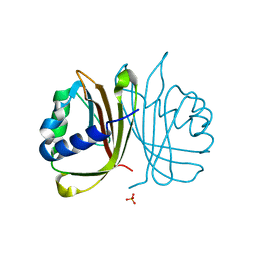 | |
3IJT
 
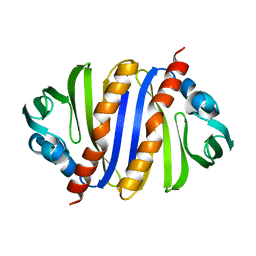 | | Structural Characterization of SMU.440, a Hypothetical Protein from Streptococcus mutans | | Descriptor: | Putative uncharacterized protein | | Authors: | Nan, J, Brostromer, E, Kristensen, O, Su, X.-D. | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance
Plos One, 4, 2009
|
|
3C9N
 
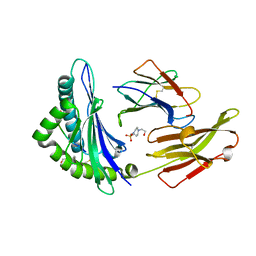 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
1FEU
 
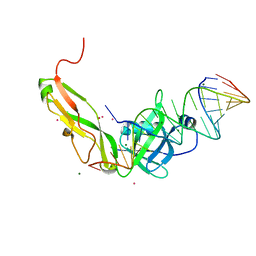 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FNM
 
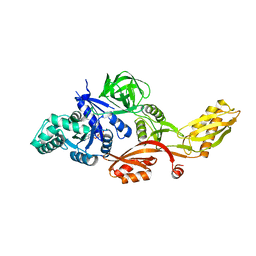 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
3UJM
 
 | |
