6DNC
 
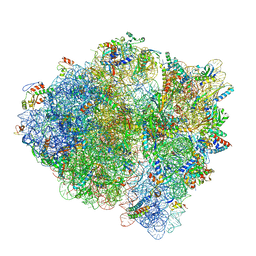 | | E.coli RF1 bound to E.coli 70S ribosome in response to UAU sense A-site codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Korostelev, A.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of premature translation termination on a sense codon.
J. Biol. Chem., 293, 2018
|
|
5D8B
 
 | |
3FBV
 
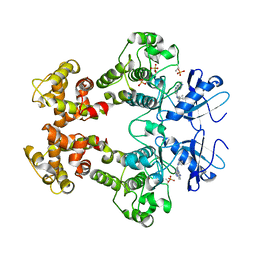 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
5J4D
 
 | |
5JUS
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure III (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5JUP
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
5JUU
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
4V9Q
 
 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | Deposit date: | 2013-06-12 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7C
 
 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7D
 
 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA*EF-G structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
8DFV
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa | | Descriptor: | CALCIUM ION, Endoribonuclease Dcr-1, Loquacious, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-22 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGI
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGA
 
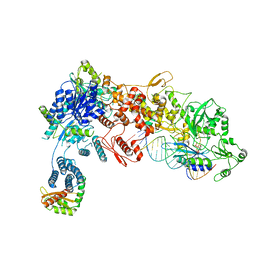 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG7
 
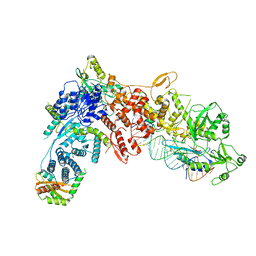 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG5
 
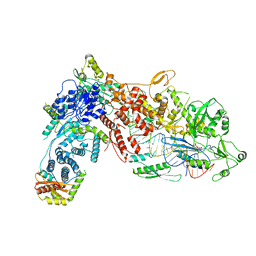 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGJ
 
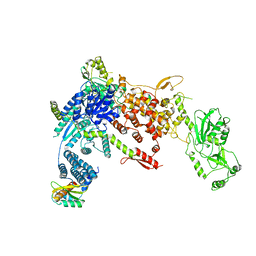 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
6OFX
 
 | | Non-rotated ribosome (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OG7
 
 | | 70S termination complex with RF2 bound to the UGA codon. Non-rotated ribosome with RF2 bound (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-01 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OGG
 
 | | 70S termination complex with RF2 bound to the UGA codon. Rotated ribosome with RF2 bound (Structure IV). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OGF
 
 | | 70S termination complex with RF2 bound to the UGA codon. Partially rotated ribosome with RF2 bound (Structure III). | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6OGI
 
 | | 70S termination complex with RF2 bound to the UAG codon. Rotated ribosome conformation (Structure V) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Demo, G, Loveland, A.B, Xu, C, Korostelev, A.A. | | Deposit date: | 2019-04-02 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Extensive ribosome and RF2 rearrangements during translation termination.
Elife, 8, 2019
|
|
6AWB
 
 | | Structure of 30S ribosomal subunit and RNA polymerase complex in non-rotated state | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6AWD
 
 | | Structure of 30S (S1 depleted) ribosomal subunit and RNA polymerase complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demo, G, Rasouly, A, Vasilyev, N, Loveland, A.B, Diaz-Avalos, R, Grigorieff, N, Nudler, E, Korostelev, A.A. | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of RNA polymerase bound to ribosomal 30S subunit.
Elife, 6, 2017
|
|
6B4V
 
 | |
