1EXM
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | Descriptor: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2000-05-03 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
5IDK
 
 | |
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
1HA3
 
 | | ELONGATION FACTOR TU IN COMPLEX WITH aurodox | | Descriptor: | BETA-MERCAPTOETHANOL, ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vogeley, L, Palm, G.J, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Change of Elongation Factor TU Induced by Antibiotic Binding: Crystal Structure of the Complex between EF-TU:Gdp and Aurodox
J.Biol.Chem., 276, 2001
|
|
2Q8W
 
 | |
2J98
 
 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
4D50
 
 | | Structure of human deoxyhypusine hydroxylase | | Descriptor: | DEOXYHYPUSINE HYDROXYLASE, FE (III) ION, GUANIDINE, ... | | Authors: | Han, Z, Sakai, N, Hilgenfeld, R. | | Deposit date: | 2014-10-31 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Peroxo-Diiron(III) Intermediate of Deoxyhypusine Hydroxylase, an Oxygenase Involved in Hypusination.
Structure, 23, 2015
|
|
2VB0
 
 | | Crystal structure of coxsackievirus B3 proteinase 3C | | Descriptor: | CHLORIDE ION, POLYPROTEIN 3BCD | | Authors: | Anand, K, Mesters, J.R, Goerlach, R, Zell, R, Hilgenfeld, R. | | Deposit date: | 2007-09-05 | | Release date: | 2008-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Coxsackie Virus B3 Proteinase 3C
To be Published
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2JBK
 
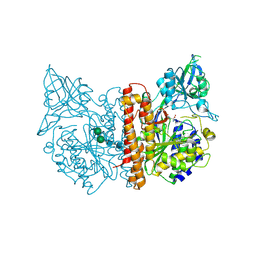 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with quisqualic acid (quisqualate, alpha-amino-3,5-dioxo-1,2,4- oxadiazolidine-2-propanoic acid) | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JBJ
 
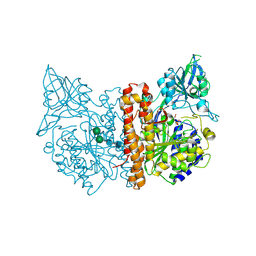 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with 2-PMPA (2-phosphonoMethyl-pentanedioic acid) | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1LK9
 
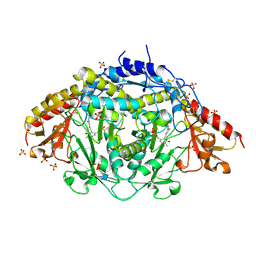 | | The Three-dimensional Structure of Alliinase from Garlic | | Descriptor: | 2-AMINO-ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kuettner, E.B, Hilgenfeld, R, Weiss, M.S. | | Deposit date: | 2002-04-24 | | Release date: | 2002-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The active principle of garlic at atomic resolution
J.Biol.Chem., 277, 2002
|
|
6Z2E
 
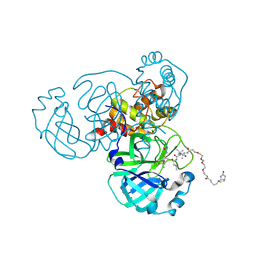 | | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone | | Descriptor: | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone
To Be Published
|
|
5LC0
 
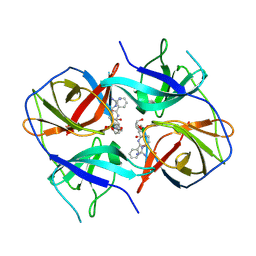 | | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor | | Descriptor: | N-((S)-3-(4-(aminomethyl)phenyl)-1-(((R)-4-guanidino-1-(5-hydroxy-1,3,2-dioxaborinan-2-yl)butyl)amino)-1-oxopropan-2-yl)benzamide, NS2B-NS3 protease,NS2B-NS3 protease | | Authors: | Lei, J, Hansen, G, Zhang, L.L, Hilgenfeld, R. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor.
Science, 353, 2016
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AIZ
 
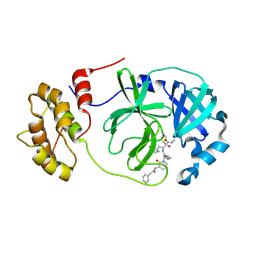 | | Mpro of SARS-CoV-2 in complex with the RK-68 inhibitor | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide, CHLORIDE ION, Replicase polyprotein 1ab | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
8AJ1
 
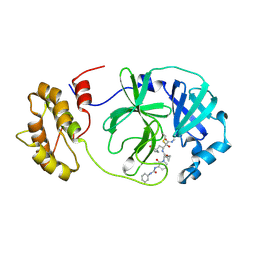 | | SARS-CoV-2 Mpro in Complex with RK-107 | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(phenylcarbamoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2024-09-11 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 2025
|
|
2V6N
 
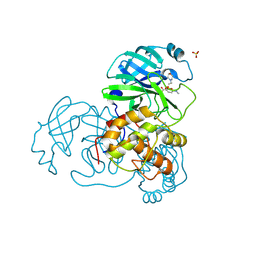 | | Crystal structures of the SARS-coronavirus main proteinase inactivated by benzotriazole compounds | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(DIMETHYLAMINO)BENZOIC ACID, REPLICASE POLYPROTEIN 1AB, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-07-19 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
2VJ1
 
 | | A Structural View of the Inactivation of the SARS-Coronavirus Main Proteinase by Benzotriazole Esters | | Descriptor: | 4-(DIMETHYLAMINO)BENZOIC ACID, BENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Verschueren, K.H.G, Pumpor, K, Anemueller, S, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Structural View of the Inactivation of the Sars Coronavirus Main Proteinase by Benzotriazole Esters.
Chem.Biol., 15, 2008
|
|
7QL8
 
 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
7QUW
 
 | | CVB3-3Cpro in complex with inhibitor MG-78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
