2GEJ
 
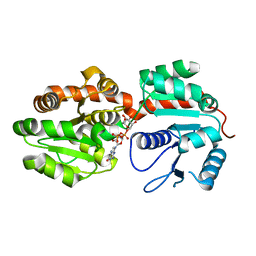 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
2GEK
 
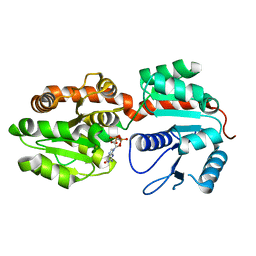 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
8QOY
 
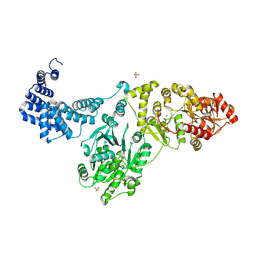 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization.
Nat.Chem.Biol., 2024
|
|
5MNI
 
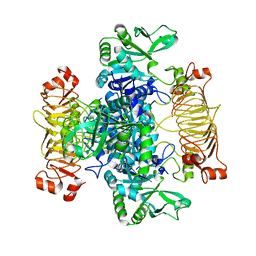 | | Escherichia coli AGPase mutant R130A apo form | | Descriptor: | Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, Marina, A, Orrantia, A, Eguskiza, A, Guerin, M.E. | | Deposit date: | 2016-12-13 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanistic insights into the allosteric regulation of bacterial ADP-glucose pyrophosphorylases.
J. Biol. Chem., 292, 2017
|
|
4Y6U
 
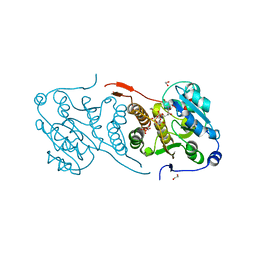 | | Mycobacterial protein | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, CHLORIDE ION, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
6MDS
 
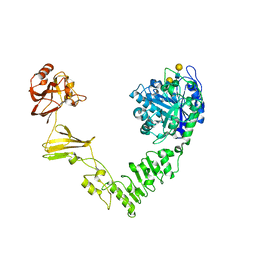 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with complex biantennary glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6MDV
 
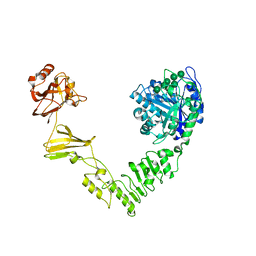 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with high-mannose glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
7NWF
 
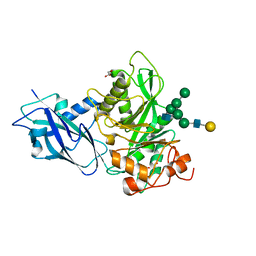 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with hybrid-type glycan (GalGlcNAcMan5GlcNAc) product | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Garcia-Alija, M, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GH18 endo-beta-N-acetylglucosaminidases use distinct mechanisms to process hybrid-type N-linked glycans.
J.Biol.Chem., 297, 2021
|
|
7OJT
 
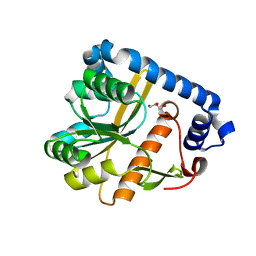 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
6H8L
 
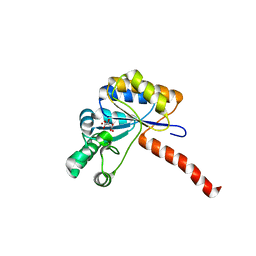 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ZINC ION | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
6H8N
 
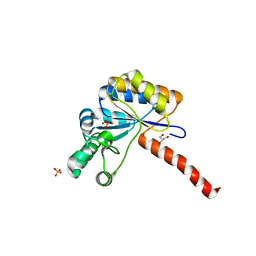 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis - mutant D285S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ... | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
6Z2D
 
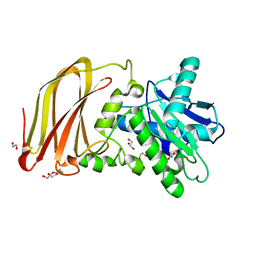 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 41 21 2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-15 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2O
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, O-glycan protease, ZINC ION | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2P
 
 | | Crystal structure of catalytic inactive OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (glycodrosocin) substrate | | Descriptor: | CALCIUM ION, Glycodrosocin, O-glycan protease, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
6Z2Q
 
 | | Crystal structure of wild type OgpA from Akkermansia muciniphila in complex with an O-glycopeptide (GalGalNAc-TS) product | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Glycodrosocin, ... | | Authors: | Trastoy, B, Naegali, A, Anso, I, Sjogren, J, Guerin, M.E. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Structural basis of mammalian mucin processing by the human gut O-glycopeptidase OgpA from Akkermansia muciniphila.
Nat Commun, 11, 2020
|
|
5NRB
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-1 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, N-acetyllactosaminide alpha-1,3-galactosyltransferase, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5OCE
 
 | | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, [(2~{R},3~{S},4~{S},5~{S},6~{S})-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hexadecanoate, ... | | Authors: | Albesa-Jove, D, Tersa, M, Guerin, M.E. | | Deposit date: | 2017-06-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria.
ACS Chem. Biol., 13, 2018
|
|
5NRE
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - a3GalT in complex with lactose - a3GalT-LAT | | Descriptor: | N-acetyllactosaminide alpha-1,3-galactosyltransferase, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NR9
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | Descriptor: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NRD
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5F31
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with palmitate - P 42 21 2 space group | | Descriptor: | ETHANOL, PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, ... | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
5F2Z
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with palmitate - P21 space group | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
5F2T
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with palmitate - C 2 space group | | Descriptor: | MAGNESIUM ION, PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
5F34
 
 | | Crystal structure of membrane associated PatA from Mycobacterium smegmatis in complex with S-hexadecyl Coenzyme A - P21 space group | | Descriptor: | Phosphatidylinositol mannoside acyltransferase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3~{S})-4-[[3-(2-hexadecylsulfanylethylamino)-3-oxidanylidene-propyl]amino]-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-butyl] hydrogen phosphate | | Authors: | Albesa-Jove, D, Svetlikova, Z, Carreras-Gonzalez, A, Tersa, M, Sancho-Vaello, E, Cifuente, J.O, Mikusova, K, Guerin, M.E. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA.
Nat Commun, 7, 2016
|
|
