1C5G
 
 | | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Goldsmith, E.J. | | Deposit date: | 1999-12-07 | | Release date: | 1999-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering of plasminogen activator inhibitor-1 to reduce the rate of latency transition.
Nat.Struct.Biol., 2, 1995
|
|
7UOS
 
 | | Structure of WNK1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 4-[bromo(dichloro)methanesulfonyl]-N-cyclohexyl-2-nitroaniline, Serine/threonine-protein kinase WNK1 | | Authors: | Akella, R, Goldsmith, E.J, Akella, R. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of WNK1 inhibitor complex
To Be Published
|
|
1GOL
 
 | |
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
8EDH
 
 | |
2ERK
 
 | | PHOSPHORYLATED MAP KINASE ERK2 | | Descriptor: | EXTRACELLULAR SIGNAL-REGULATED KINASE 2 | | Authors: | Canagarajah, B.J, Goldsmith, E.J. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation mechanism of the MAP kinase ERK2 by dual phosphorylation.
Cell(Cambridge,Mass.), 90, 1997
|
|
6OT6
 
 | | Rat ERK2 D319N | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Taylor, C.A, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional divergence caused by mutations in an energetic hotspot in ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OL2
 
 | | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high-throughput-screening | | Descriptor: | ACETATE ION, GLYCEROL, N-[2-(5,8-dimethoxy-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]-2-iodobenzamide, ... | | Authors: | Chlebowicz, J, Akella, R, Sekulski, K, Humphreys, J.M, Durbacz, M.Z, He, H, Rodan, A, Posner, B, Goldsmith, E.J. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallography of novel WNK1 and WNK3 inhibitors discovered from high throughput screening
To Be Published
|
|
6OTS
 
 | | Rat ERK2 E320K | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Taylor, C.A, Cormier, K.W, Juang, Y.-C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2019-05-03 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional divergence caused by mutations in an energetic hotspot in ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL6
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL7
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
5D9H
 
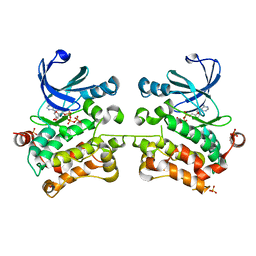 | | Crystal structure of SPAK (STK39) dimer in the basal activity state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, STE20/SPS1-related proline-alanine-rich protein kinase, ... | | Authors: | Taylor, C.A, Juang, Y.C, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain-Swapping Switch Point in Ste20 Protein Kinase SPAK.
Biochemistry, 54, 2015
|
|
5UOJ
 
 | |
5UMO
 
 | |
3DAK
 
 | |
3ENM
 
 | | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 6, GLYCEROL, ... | | Authors: | Min, X, Akella, R, He, H, Humphreys, J.M, Tsutakawa, S, Lee, S.-J, Tainer, J.A, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer
Structure, 17, 2009
|
|
5W7T
 
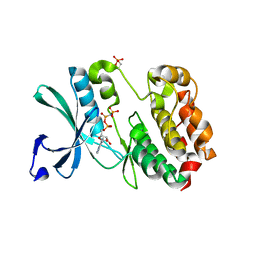 | |
3ERK
 
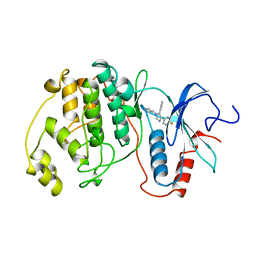 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
2GCD
 
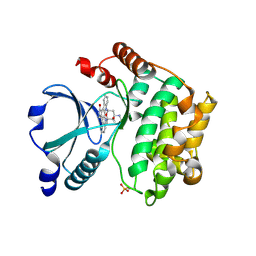 | | TAO2 kinase domain-staurosporine structure | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase TAO2 | | Authors: | Zhou, T, Sun, L, Gao, Y, Earnest, S, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2006-03-14 | | Release date: | 2006-09-05 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the MAP3K TAO2 kinase domain bound by an inhibitor staurosporine.
Acta Biochim.Biophys.Sinica, 38, 2006
|
|
2GPH
 
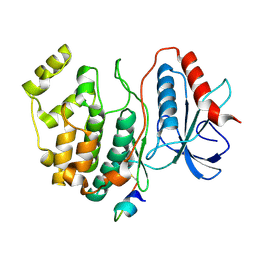 | | Docking motif interactions in the MAP kinase ERK2 | | Descriptor: | Mitogen-activated protein kinase 1, Tyrosine-protein phosphatase non-receptor type 7 | | Authors: | Zhou, T, Sun, L, Humphreys, J, Goldsmith, E.J. | | Deposit date: | 2006-04-17 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Docking Interactions Induce Exposure of Activation Loop in the MAP Kinase ERK2.
Structure, 14, 2006
|
|
2LRU
 
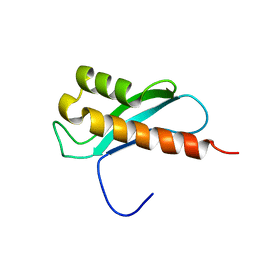 | | Solution Structure of the WNK1 Autoinhibitory Domain | | Descriptor: | Serine/threonine-protein kinase WNK1 | | Authors: | Moon, T.M, Correa, F, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the WNK1 Autoinhibitory Domain, a WNK-Specific PF2 Domain.
J.Mol.Biol., 425, 2013
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
3P4K
 
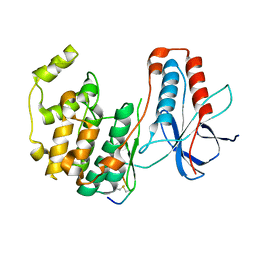 | | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution | | Descriptor: | MAP kinase 14, Mitogen-activated protein kinase 14 | | Authors: | Akella, R, Min, X, Wu, Q, Gardner, K.H, Goldsmith, E.J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | The third conformation of p38a MAP kinase observed in phosphorylated p38a and in solution
Structure, 18, 2010
|
|
