2GCZ
 
 | | Solution Structure of alpha-Conotoxin OmIA | | Descriptor: | Alpha-conotoxin OmIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-03-15 | | Release date: | 2006-07-25 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist alpha-conotoxin OmIA that discriminates alpha3 vs. alpha6 nAChR subtypes
Biochem.Biophys.Res.Commun., 345, 2006
|
|
2I28
 
 | | Solution Structure of alpha-Conotoxin BuIA | | Descriptor: | Alpha-conotoxin BuIA | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of alpha-conotoxin BuIA, a novel neuronal nicotinic acetylcholine receptor antagonist with an unusual 4/4 disulfide scaffold
Biochem.Biophys.Res.Commun., 349, 2006
|
|
2G9L
 
 | |
1PQR
 
 | | Solution Conformation of alphaA-Conotoxin EIVA | | Descriptor: | Alpha-A-conotoxin EIVA | | Authors: | Chi, S.-W, Park, K.-H, Suk, J.-E, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Conformation of alphaA-conotoxin EIVA, a Potent Neuromuscular Nicotinic Acetylcholine Receptor Antagonist from Conus ermineus
J.Biol.Chem., 278, 2003
|
|
1UL2
 
 | | Solution Conformation of alpha-Conotoxin GIC | | Descriptor: | alpha-conotoxin GIC | | Authors: | Chi, S.-W, Kim, D.-H, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2003-09-06 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin GIC, a novel potent antagonist of alpha3beta2 nicotinic acetylcholine receptors
Biochem.J., 380, 2004
|
|
1ZLC
 
 | | Solution Conformation of alpha-conotoxin PIA | | Descriptor: | Alpha-conotoxin PIA | | Authors: | Chi, S.-W, Lee, S.-H, Kim, D.-H, Kim, J.-S, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | Deposit date: | 2005-05-05 | | Release date: | 2006-05-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha-conotoxin PIA, a novel antagonist of alpha6 subunit containing nicotinic acetylcholine receptors
Biochem.Biophys.Res.Commun., 338, 2005
|
|
2EH7
 
 | | Crystal structure of humanized KR127 FAB | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EH8
 
 | | Crystal structure of the complex of humanized KR127 fab and PRES1 peptide epitope | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6IJQ
 
 | | Solution structure of BCL-XL bound to P73-TAD peptide | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Tumor protein p73 | | Authors: | Yoon, M.-K, Ha, J.-H, Lee, M.-S, Chi, S.-W. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cytoplasmic pro-apoptotic function of the tumor suppressor p73 is mediated through a modified mode of recognition of the anti-apoptotic regulator Bcl-XL.
J. Biol. Chem., 293, 2018
|
|
4B04
 
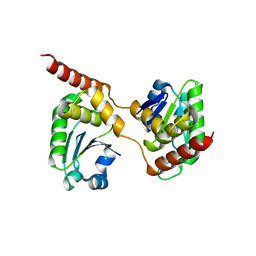 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3M7M
 
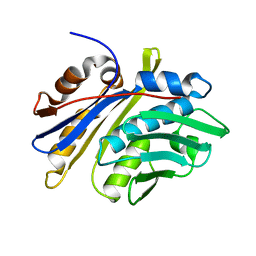 | | Crystal structure of monomeric hsp33 | | Descriptor: | 33 kDa chaperonin | | Authors: | Chi, S.W, Jeong, D.G, Woo, J.R, Park, B.C, Ryu, S.E, Kim, S.J. | | Deposit date: | 2010-03-16 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of monomeric hsp33
To be Published
|
|
1COK
 
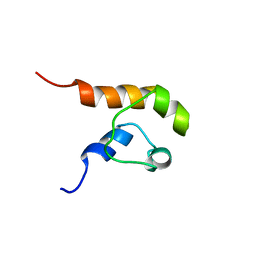 | |
7BOL
 
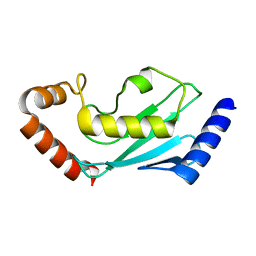 | | ubiquitin-conjugating enzyme, Ube2D2 | | Descriptor: | Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
1I7F
 
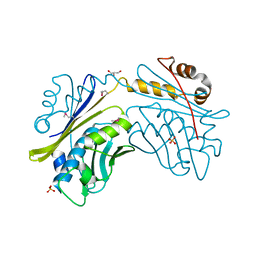 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | Authors: | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-05-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
1KU0
 
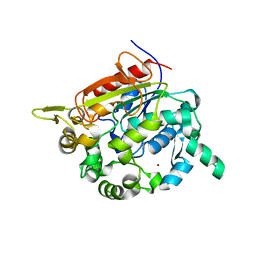 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
5GTJ
 
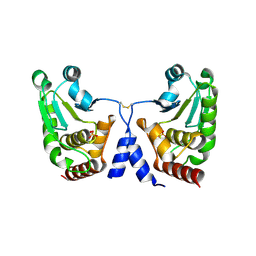 | |
6M2D
 
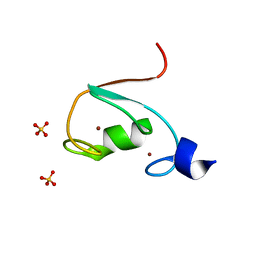 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
 | |
1DXS
 
 | |
