8IU7
 
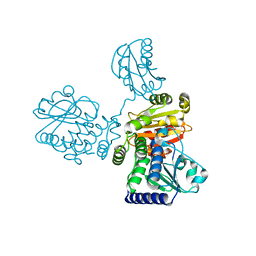 | |
6SER
 
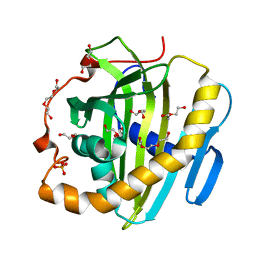 | | Crystal structure of human STARD10 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, START domain-containing protein 10, ... | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | The crystal structure of human STARD10
To Be Published
|
|
6SJF
 
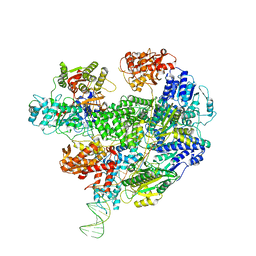 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJG
 
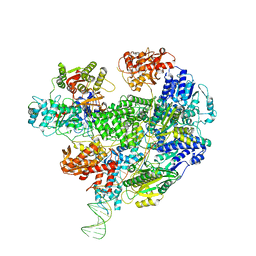 | | Cryo-EM structure of the RecBCD no Chi negative control complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJB
 
 | | Cryo-EM structure of the RecBCD Chi recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJE
 
 | | Cryo-EM structure of the RecBCD Chi partially-recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T2U
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-minus2 substrate | | Descriptor: | DNA (Chi-minus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T2V
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-plus2 substrate | | Descriptor: | DNA (Chi-plus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2020-01-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6FWS
 
 | |
6FWR
 
 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
8IOO
 
 | |
7YKM
 
 | |
6LRD
 
 | | Structure of RecJ complexed with a 5'-P-dSpacer-modified ssDNA | | Descriptor: | ASP-LEU-PRO-PHE, DNA (5'-D(P*(3DR)P*TP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Cheng, K, Hua, Y. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901335 Å) | | Cite: | Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans.
Nucleic Acids Res., 48, 2020
|
|
7W89
 
 | | The structure of Deinococcus radiodurans Yqgf | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Cheng, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.500061 Å) | | Cite: | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
7W8D
 
 | | The structure of Deinococcus radiodurans RuvC | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75016141 Å) | | Cite: | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
5F54
 
 | | Structure of RecJ complexed with dTMP | | Descriptor: | MANGANESE (II) ION, Single-stranded-DNA-specific exonuclease, THYMIDINE-5'-PHOSPHATE | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5F56
 
 | | Structure of RecJ complexed with DNA and SSB-ct | | Descriptor: | ALA-ASP-LEU-PRO-PHE, DNA (5'-D(*CP*TP*GP*AP*TP*GP*GP*CP*A)-3'), MANGANESE (II) ION, ... | | Authors: | Zhao, Y, Hua, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5F55
 
 | | Structure of RecJ complexed with DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*C)-3'), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
7WRW
 
 | | Structure of Deinococcus radiodurans HerA | | Descriptor: | HerA | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.00008273 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
7WRX
 
 | | Structure of Deinococcus radiodurans HerA-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HerA, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.40003562 Å) | | Cite: | Structural and DNA end resection study of the bacterial NurA-HerA complex.
Bmc Biol., 21, 2023
|
|
5GGM
 
 | | The NMR structure of calmodulin in CTAB reverse micelles | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Xu, G, Cheng, K, Wu, Q, Liu, M, Li, C. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of calmodulin in CTAB reverse micelles
To Be Published
|
|
4HQP
 
 | | Alpha7 nicotinic receptor chimera and its complex with Alpha bungarotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Li, S.X, Cheng, K, Gomoto, R, Bren, N, Huang, S, Sine, S, Chen, L. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural principles for Alpha-neurotoxin binding to and selectivity among nicotinic receptors
To be Published
|
|
1Q4K
 
 | | The polo-box domain of Plk1 in complex with a phospho-peptide | | Descriptor: | Phospho-peptide sequence Met.Gln.Ser.pThr.Pro.Leu, Serine/threonine-protein kinase PLK | | Authors: | Cheng, K, Lowe, E.D, Sinclair, J, Nigg, E.A, Johnson, L.N. | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex.
Embo J., 22, 2003
|
|
7SH0
 
 | | CRYSTAL STRUCTURE OF ENDOPLASMIC RETICULUM AMINOPEPTIDASE 2 (ERAP2) COMPLEX WITH A HIGHLY SELECTIVE AND POTENT SMALL MOLECULE | | Descriptor: | (2S)-N-hydroxy-3-(4-methoxyphenyl)-2-[4-({[5-(pyridin-2-yl)thiophene-2-sulfonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Li, L, Bouvier, M. | | Deposit date: | 2021-10-07 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2QN1
 
 | | Glycogen Phosphorylase b in complex with asiatic acid | | Descriptor: | Glycogen phosphorylase, muscle form, asiatic acid | | Authors: | Zographos, S.E, Leonidas, D.D, Alexacou, K.-M, Hayes, J, Oikonomakos, N.G. | | Deposit date: | 2007-07-17 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Naturally occurring pentacyclic triterpenes as inhibitors of glycogen phosphorylase: synthesis, structure-activity relationships, and X-ray crystallographic studies
J.Med.Chem., 51, 2008
|
|
