3KH8
 
 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
8R0F
 
 | | Capsid structure of Giardiavirus (GLV) HP strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) HP strain
To Be Published
|
|
8R0G
 
 | | Capsid structure of Giardiavirus (GLV) CAT strain | | Descriptor: | Capsid protein | | Authors: | Wang, H, Gianluca, M, Munke, A, Hassan, M.M, Lalle, M, Okamoto, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Capsid structure of Giardiavirus (GLV) CAT strain
To Be Published
|
|
7OHA
 
 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHB
 
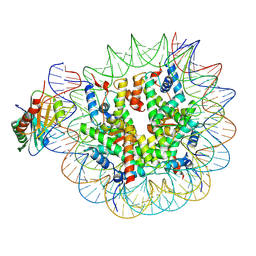 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OH9
 
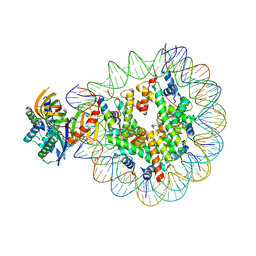 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
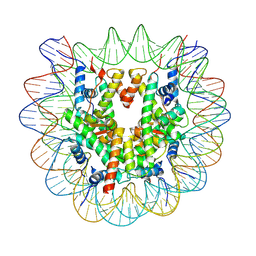 | |
7OS7
 
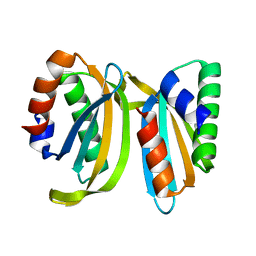 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
6IOJ
 
 | |
6IO4
 
 | |
7SCG
 
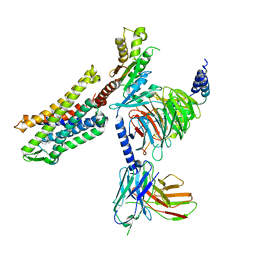 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6HBA
 
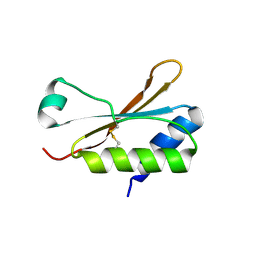 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBC
 
 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBB
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
3LFV
 
 | | crystal structure of unliganded PDE5A GAF domain | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
3MF0
 
 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
6I6I
 
 | | Circular permutant of ribosomal protein S6, adding 6aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6, SULFATE ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6S
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6E
 
 | |
6I69
 
 | |
6I6O
 
 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6Y
 
 | |
6I6U
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to N terminal of P81-82, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I6W
 
 | |
