3NMP
 
 | | Crystal structure of the abscisic receptor PYL2 mutant A93F in complex with pyrabactin | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMT
 
 | | Crystal structure of pyrabactin bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase HAB1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMV
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL2 mutant A93F in complex with type 2C protein phosphatase ABI2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NMN
 
 | | Crystal structure of pyrabactin-bound abscisic acid receptor PYL1 in complex with type 2C protein phosphatase ABI1 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KAY
 
 | | Crystal structure of abscisic acid receptor PYL1 | | Descriptor: | Putative uncharacterized protein At5g46790 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB0
 
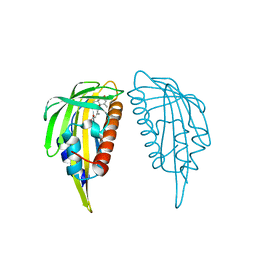 | | Crystal structure of abscisic acid-bound PYL2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KAZ
 
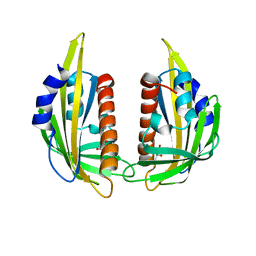 | | Crystal structure of abscisic acid receptor PYL2 | | Descriptor: | 1,3-BUTANEDIOL, Putative uncharacterized protein At2g26040 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3KB3
 
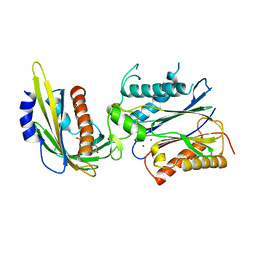 | | Crystal structure of abscisic acid-bound PYL2 in complex with HAB1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Melcher, K, Soon, F.-F, Ng, L.-M, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Xu, H.E. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Agate-latch-lock mechanism for hormone signalling by abscisic acid receptors
Nature, 462, 2009
|
|
3NMH
 
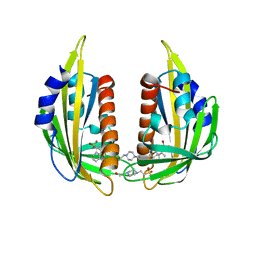 | | Crystal structure of the abscisic receptor PYL2 in complex with pyrabactin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Zhou, X.E, Melcher, K, Ng, L.-M, Soon, F.-F, Xu, Y, Suino-Powell, K.M, Kovach, A, Li, J, Yong, E.-L, Xu, H.E. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and mechanism of ABA receptor antagonism.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7S3F
 
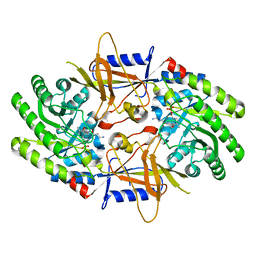 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
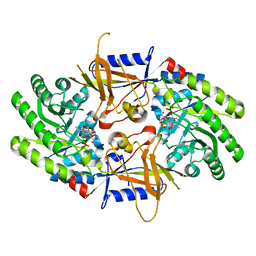 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
9B8N
 
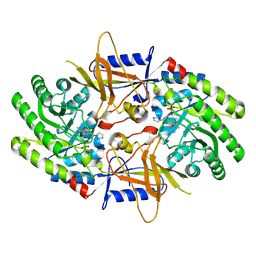 | | Crystal structure of ornithine decarboxylase in complex with a novel inhibitor (10-S) | | Descriptor: | O-{[(3S)-pyrrolidin-3-yl]methyl}hydroxylamine, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schultz, C.R, Aleiwi, B, Zhou, X.E, Suino-Powell, K, Melcher, K, Brunzelle, J.S, Almeida, N.M.S, Wilson, A.K, Ellsworth, E, Bachmann, A.S. | | Deposit date: | 2024-03-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Biological Activity of Novel Ornithine Decarboxylase (ODC) Inhibitors.
J.Med.Chem., 68, 2025
|
|
9B8M
 
 | | Crystal structure of ornithine decarboxylase in complex with a novel inhibitor | | Descriptor: | O-{[(3R)-pyrrolidin-3-yl]methyl}hydroxylamine, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schultz, C.R, Aleiwi, B, Zhou, X.E, Suino-Powell, K, Melcher, K, Brunzelle, J.S, Almeida, N.M.S, Wilson, A.K, Ellsworth, E, Bachmann, A.S. | | Deposit date: | 2024-03-31 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Activity of Novel Ornithine Decarboxylase (ODC) Inhibitors.
J.Med.Chem., 68, 2025
|
|
6VN7
 
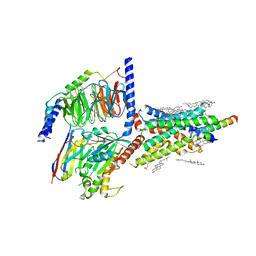 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
6PB0
 
 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6PA7
 
 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
6C9H
 
 | | non-phosphorylated AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
7U6U
 
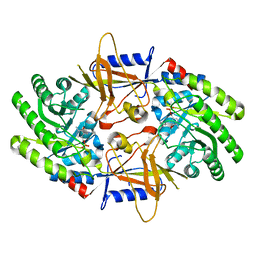 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R in complex with PLP | | Descriptor: | Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
7U6P
 
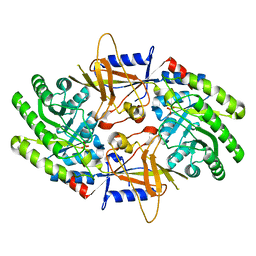 | | Structure of an intellectual disability-associated ornithine decarboxylase variant G84R | | Descriptor: | Ornithine decarboxylase, PHOSPHATE ION | | Authors: | Zhou, X.E, Schultz, C.R, Powell, K.S, Henrickson, A, Lamp, J, Brunzelle, J.S, Demeler, B, Vega, I.E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Enzymatic Activity of an Intellectual Disability-Associated Ornithine Decarboxylase Variant, G84R.
Acs Omega, 7, 2022
|
|
5W0P
 
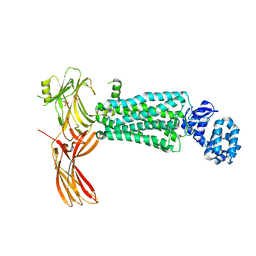 | | Crystal structure of rhodopsin bound to visual arrestin determined by X-ray free electron laser | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, He, Y, de Waal, P.W, Gao, X, Kang, Y, Van Eps, N, Yin, Y, Pal, K, Goswami, D, White, T.A, Barty, A, Latorraca, N.R, Chapman, H.N, Hubbell, W.L, Dror, R.O, Stevens, R.C, Cherezov, V, Gurevich, V.V, Griffin, P.R, Ernst, O.P, Melcher, K, Xu, H.E. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors.
Cell, 170, 2017
|
|
5JA5
 
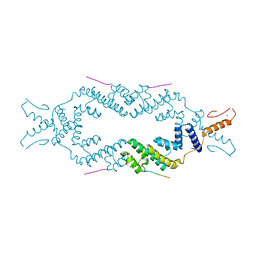 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5J9K
 
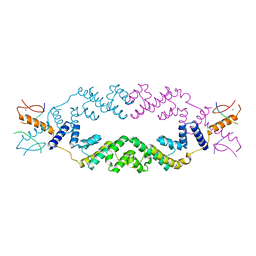 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, ZINC ION, rice D53 peptide 794-808 | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
