6U0R
 
 | |
6U0V
 
 | |
1P5Y
 
 | |
1P5W
 
 | |
3RA9
 
 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*A)-3') | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
3RA8
 
 | | Structural studies of AAV8 capsid transitions associated with endosomal trafficking | | Descriptor: | ADENOSINE MONOPHOSPHATE, Capsid protein | | Authors: | Nam, H.-J, Gurda, B, McKenna, R, Porter, M, Byrne, B, Salganik, M, Muzyczka, N, Agbandje-McKenna, M. | | Deposit date: | 2011-03-27 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking.
J.Virol., 85, 2011
|
|
2ANL
 
 | | X-ray crystal structure of the aspartic protease plasmepsin 4 from the malarial parasite plasmodium malariae bound to an allophenylnorstatine based inhibitor | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, plasmepsin IV | | Authors: | Clemente, J.C, Govindasamy, L, Madabushi, A, Fisher, S.Z, Moose, R.E, Yowell, C.A, Hidaka, K, Kimura, T, Hayashi, Y, Kiso, Y, Agbandje-McKenna, M, Dame, J.B, Dunn, B.M, McKenna, R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5JJ8
 
 | | Crystal Structure of the Beta Carbonic Anhydrase psCA3 isolated from Pseudomonas aeruginosa - alternate crystal packing form | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Pinard, M.A, Kurian, J.J, Aggarwal, M, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Cryoannealing-induced space-group transition of crystals of the carbonic anhydrase psCA3.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6NXE
 
 | |
6B9Q
 
 | |
6O9R
 
 | |
4FIK
 
 | | Human carbonic anhydrase II H64A complexed with thioxolone hydrolysis products | | Descriptor: | 2,4-dihydroxybenzenesulfenic acid, 4,4'-disulfanediyldibenzene-1,3-diol, 4-MERCAPTOBENZENE-1,3-DIOL, ... | | Authors: | Sippel, K.H, Genis, C, Govindasamy, L, Agbandje-McKenna, M, Tripp, B.C, McKenna, R. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synchrotron Radiation Provides a Plausible Explanation for the Generation of a Free Radical Adduct of Thioxolone in Mutant Carbonic Anhydrase II.
J Phys Chem Lett, 1, 2010
|
|
5URF
 
 | | The structure of human bocavirus 1 | | Descriptor: | viral protein 3 | | Authors: | Mietzsch, M, Kailasan, S, Garrison, J, Ilyas, M, Chipman, P, Kandola, K, Jansen, M, Spear, J, Sousa, D, McKenna, R, Soderlund-Venermo, M, Baker, T, Agbandje-McKenna, M. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Insights into Human Bocaparvoviruses.
J. Virol., 91, 2017
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
5IPK
 
 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
7L0X
 
 | |
7L0Y
 
 | |
4IOV
 
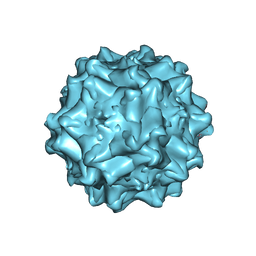 | | The structure of AAVrh32.33, a Novel Gene Delivery Vector | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Capsid protein VP1 | | Authors: | Mikalism, K, Nam, H.-J, Van vilet, K, Vandenberghe, L.H, Mays, L.E, McKenna, R, Wilson, J, Agbandje-McKenna, M. | | Deposit date: | 2013-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of AAVrh32.33, a novel gene delivery vector.
J.Struct.Biol., 186, 2014
|
|
5US9
 
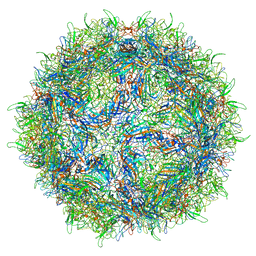 | | Human bocavirus 4 | | Descriptor: | Capsid protein VP2 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into Human Bocaparvoviruses.
J. Virol., 91, 2017
|
|
5US7
 
 | | Human bocavirus 3 | | Descriptor: | VP2 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural Insights into Human Bocaparvoviruses.
J. Virol., 91, 2017
|
|
1Z1C
 
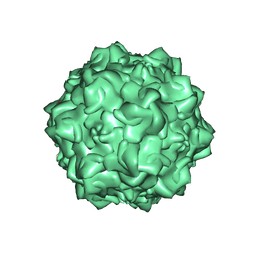 | | Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute virus of Mice | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*AP*CP*AP*CP*CP*AP*AP*AP*A)-3', 5'-D(*AP*TP*CP*CP*TP*CP*TP*AP*TP*CP*AP*C)-3', ... | | Authors: | Kontou, M, Govindasamy, L, Nam, H.J, Bryant, N, Llamas-Saiz, A.L, Foces-Foces, C, Hernando, E, Rubio, M.P, McKenna, R, Almendral, J.M, Agbandje-McKenna, M. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice.
J.Virol., 79, 2005
|
|
1MVM
 
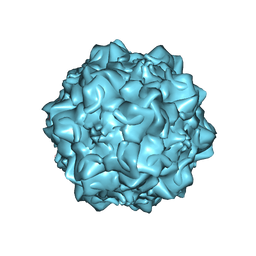 | | MVM(STRAIN I), COMPLEX(VIRAL COAT/DNA), VP2, PH=7.5, T=4 DEGREES C | | Descriptor: | DNA (5'-D(*CP*AP*AP*A)-3'), DNA (5'-D(*CP*CP*AP*CP*CP*CP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*A)-3'), ... | | Authors: | Llamas-Saiz, A.L, Agbandje-McKenna, M, Rossmann, M.G. | | Deposit date: | 1996-06-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure determination of minute virus of mice.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1MOO
 
 | | Site Specific Mutant (H64A) of Human Carbonic Anhydrase II at high resolution | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic Anhydrase II, MERCURY (II) ION, ... | | Authors: | Duda, D.M, Govindasamy, L, Agbandje-McKenna, M, Tu, C.K, Silverman, D.N, McKenna, R. | | Deposit date: | 2002-09-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The refined atomic structure of carbonic anhydrase II at 1.05 A resolution: implications of chemical rescue of proton transfer.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4QC8
 
 | | Structural annotation of pathogenic bovine Parvovirus-1 | | Descriptor: | VP2 | | Authors: | Kailasan, S, Halder, S, Gurda, B.L, Bladek, H, Chipman, P.R, McKenna, R, Brown, K, Agbandje-McKenna, M. | | Deposit date: | 2014-05-09 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an enteric pathogen, bovine parvovirus.
J.Virol., 89, 2015
|
|
4GBT
 
 | | Structural characterization of H-1 Parvovirus: comparison of infectious virions to replication defective particles | | Descriptor: | CHLORIDE ION, Capsid protein VP1, SODIUM ION | | Authors: | Halder, S, Nam, H.-J, Govindasamy, L, Vogel, M, Dinsart, C, Salome, N, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2012-07-27 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural characterization of h-1 parvovirus: comparison of infectious virions to empty capsids.
J.Virol., 87, 2013
|
|
