2FAL
 
 | | X-RAY CRYSTAL STRUCTURE OF FERRIC APLYSIA LIMACINA MYOGLOBIN IN DIFFERENT LIGANDED STATES | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Conti, E, Moser, C, Rizzi, M, Mattevi, A, Lionetti, C, Coda, A, Ascenzi, P, Brunori, M, Bolognesi, M. | | Deposit date: | 1993-06-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of ferric Aplysia limacina myoglobin in different liganded states.
J.Mol.Biol., 233, 1993
|
|
2FAM
 
 | | X-RAY CRYSTAL STRUCTURE OF FERRIC APLYSIA LIMACINA MYOGLOBIN IN DIFFERENT LIGANDED STATES | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Conti, E, Moser, C, Rizzi, M, Mattevi, A, Lionetti, C, Coda, A, Ascenzi, P, Brunori, M, Bolognesi, M. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of ferric Aplysia limacina myoglobin in different liganded states.
J.Mol.Biol., 233, 1993
|
|
1BCU
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN AND PROFLAVIN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, PROFLAVIN | | Authors: | Conti, E, Rivetti, C, Wonacott, A, Brick, P. | | Deposit date: | 1998-05-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and spectrophotometric studies of the binding of proflavin to the S1 specificity pocket of human alpha-thrombin.
FEBS Lett., 425, 1998
|
|
1AMU
 
 | | PHENYLALANINE ACTIVATING DOMAIN OF GRAMICIDIN SYNTHETASE 1 IN A COMPLEX WITH AMP AND PHENYLALANINE | | Descriptor: | ADENOSINE MONOPHOSPHATE, GRAMICIDIN SYNTHETASE 1, MAGNESIUM ION, ... | | Authors: | Conti, E, Stachelhaus, T, Marahiel, M.A, Brick, P. | | Deposit date: | 1997-06-18 | | Release date: | 1998-07-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S.
EMBO J., 16, 1997
|
|
1LCI
 
 | | FIREFLY LUCIFERASE | | Descriptor: | LUCIFERASE | | Authors: | Conti, E, Franks, N.P, Brick, P. | | Deposit date: | 1996-06-01 | | Release date: | 1997-03-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes.
Structure, 4, 1996
|
|
1EE4
 
 | |
1EE5
 
 | |
1BK6
 
 | | KARYOPHERIN ALPHA (YEAST) + SV40 T ANTIGEN NLS | | Descriptor: | KARYOPHERIN ALPHA, LARGE T ANTIGEN | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BK5
 
 | | KARYOPHERIN ALPHA FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | COBALT (II) ION, KARYOPHERIN ALPHA | | Authors: | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BA3
 
 | | FIREFLY LUCIFERASE IN COMPLEX WITH BROMOFORM | | Descriptor: | LUCIFERASE, TRIBROMOMETHANE | | Authors: | Franks, N.P, Jenkins, A, Conti, E, Lieb, W.R, Brick, P. | | Deposit date: | 1998-04-21 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the inhibition of firefly luciferase by a general anesthetic.
Biophys.J., 75, 1998
|
|
2QFA
 
 | | Crystal structure of a Survivin-Borealin-INCENP core complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Baculoviral IAP repeat-containing protein 5, Borealin, ... | | Authors: | Jeyaprakash, A.A, Klein, U.R, Lindner, D, Ebert, J, Nigg, E.A, Conti, E. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Survivin-Borealin-INCENP Core Complex Reveals How Chromosomal Passengers Travel Together.
Cell(Cambridge,Mass.), 131, 2007
|
|
4CT6
 
 | | CNOT9-CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
2C39
 
 | | RNase PH core of the archaeal exosome in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, PROBABLE EXOSOME COMPLEX EXONUCLEASE 2 | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-05 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
2C37
 
 | | RNASE PH CORE OF THE ARCHAEAL EXOSOME IN COMPLEX WITH U8 RNA | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, CHLORIDE ION, PROBABLE EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of 3' End RNA Recognition and Exoribonucleolytic Cleavage by an Exosome Rnase Ph Core.
Mol.Cell, 20, 2005
|
|
8RXB
 
 | | Human UPF1 CH domain in complex with SMG6 peptide | | Descriptor: | Regulator of nonsense transcripts 1, Telomerase-binding protein EST1A, ZINC ION | | Authors: | Langer, L, Basquin, J, Conti, E. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.
Nucleic Acids Res., 52, 2024
|
|
1W3B
 
 | | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | | Descriptor: | CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE--PEPTIDE N-ACETYLGLUCOSAMINYLTRANSFERASE 110 | | Authors: | Jinek, M, Rehwinkel, J, Lazarus, B.D, Izaurralde, E, Hanover, J.A, Conti, E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Superhelical Tpr-Repeat Domain of O-Linked Glcnac Transferase Exhibits Structural Similarities to Importin Alpha
Nat.Struct.Mol.Biol., 11, 2004
|
|
6FT6
 
 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | Descriptor: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
4UM2
 
 | | Crystal structure of the TPR domain of SMG6 | | Descriptor: | GLYCEROL, TELOMERASE-BINDING PROTEIN EST1A | | Authors: | Chakrabarti, S, Bonneau, F, Schuessler, S, Eppinger, E, Conti, E. | | Deposit date: | 2014-05-14 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phospho-Dependent and Phospho-Independent Interactions of the Helicase Upf1 with the Nmd Factors Smg5-Smg7 and Smg6.
Nucleic Acids Res., 42, 2014
|
|
6SYT
 
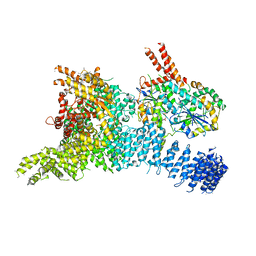 | | Structure of the SMG1-SMG8-SMG9 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gat, Y, Schuller, J.M, Conti, E. | | Deposit date: | 2019-10-01 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | InsP6binding to PIKK kinases revealed by the cryo-EM structure of an SMG1-SMG8-SMG9 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3N89
 
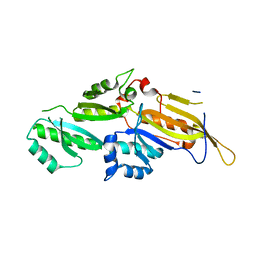 | | KH domains | | Descriptor: | Defective in germ line development protein 3, isoform a | | Authors: | Nakel, K, Hartung, S.A, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Four KH domains of the C. elegans Bicaudal-C ortholog GLD-3 form a globular structural platform.
Rna, 16, 2010
|
|
8BFI
 
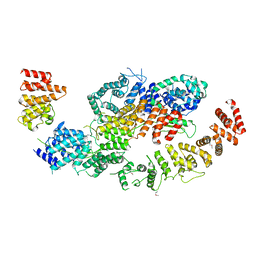 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BFH
 
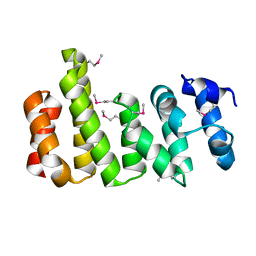 | | CNOT11 | | Descriptor: | CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BFJ
 
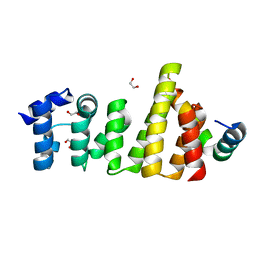 | | CNOT11-GGNBP2 complex | | Descriptor: | 1,2-ETHANEDIOL, CCR4-NOT transcription complex subunit 11, Gametogenetin-binding protein 2 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
6YVH
 
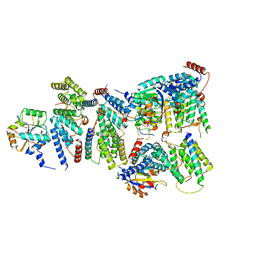 | | CWC22-CWC27-EIF4A3 Complex | | Descriptor: | Eukaryotic initiation factor 4A-III, Pre-mRNA-splicing factor CWC22 homolog, Spliceosome-associated protein CWC27 homolog | | Authors: | Basquin, J, Busetto, V, LeHir, H, Conti, E. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural and functional insights into CWC27/CWC22 heterodimer linking the exon junction complex to spliceosomes.
Nucleic Acids Res., 48, 2020
|
|
5JEA
 
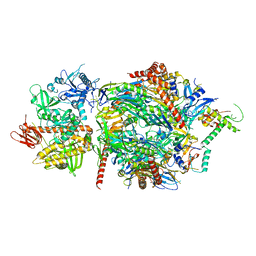 | | Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Exosome complex component CSL4, Exosome complex component MTR3, ... | | Authors: | Kowalinski, E, Ebert, J, Stegmann, E, Conti, E. | | Deposit date: | 2016-04-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Mol.Cell, 63, 2016
|
|
