2F23
 
 | |
3UJJ
 
 | |
3UJI
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2558 in complex with MN peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein gp160, ... | | Authors: | Kong, X.P. | | Deposit date: | 2011-11-07 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Human Anti-V3 HIV-1 Monoclonal Antibodies Encoded by the VH5-51/VL Lambda Genes Define a Conserved Antigenic Structure.
Plos One, 6, 2011
|
|
3GHE
 
 | |
3GO1
 
 | |
3GHB
 
 | |
1HXW
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-84538 | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Park, C.H, Nienaber, V, Kong, X.P. | | Deposit date: | 1997-01-24 | | Release date: | 1998-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
4D9L
 
 | | Fab structure of anti-HIV-1 gp120 V2 mAb 697 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697, Light chain of Fab fragment of anti-HIV1 gp120 V2 mAb 697 | | Authors: | Pan, R.M, Kong, X.P. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals.
Virology, 427, 2012
|
|
7RAI
 
 | |
1PRO
 
 | | HIV-1 PROTEASE DIMER COMPLEXED WITH A-98881 | | Descriptor: | (5R,6R)-2,4-BIS-(4-HYDROXY-3-METHOXYBENZYL)-1,5-DIBENZYL-3-OXO-6-HYDROXY-1,2,4-TRIAZACYCLOHEPTANE, HIV-1 PROTEASE | | Authors: | Park, C.H, Kong, X.P, Dealwis, C.G. | | Deposit date: | 1995-07-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel, picomolar inhibitor of human immunodeficiency virus type 1 protease.
J.Med.Chem., 39, 1996
|
|
8TR3
 
 | | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CNE40 SOSIP Envelope glycoprotein gp120, ... | | Authors: | Chan, K.-W, Kong, X.P. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains.
Nat Commun, 15, 2024
|
|
3Q6G
 
 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
3Q6F
 
 | | Crystal structure of Fab of human mAb 2909 specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of Fab of human mAb 2909, Light chain of Fab of human mAb 2909 | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.192 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
3RQW
 
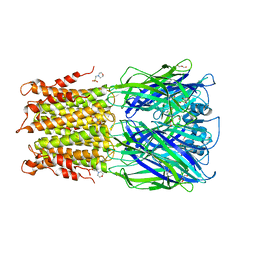 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3RQU
 
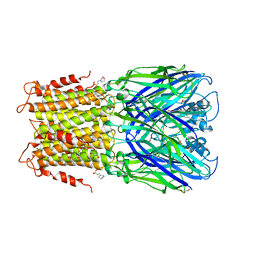 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
4M1D
 
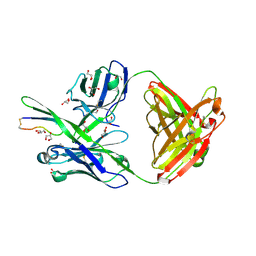 | |
4OAW
 
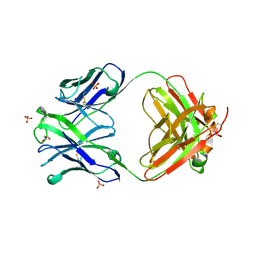 | | Fab structure of anti-HIV gp120 V2 mAb 2158 | | Descriptor: | GLYCEROL, Heavy chain of Fab fragment of anti-HIV1 gp120 V2 mAb 2158, Light chain of Fab fragment of anti-HIV1 gp120 V2 mAb 2158, ... | | Authors: | Spurrier, B.R, Kong, X.P. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional Implications of the Binding Mode of a Human Conformation-Dependent V2 Monoclonal Antibody against HIV.
J.Virol., 88, 2014
|
|
4F57
 
 | |
4F58
 
 | |
4F8H
 
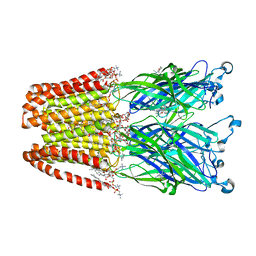 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
6VY2
 
 | |
4JO3
 
 | |
4JO4
 
 | | Crystal structure of rabbit mAb R20 Fab | | Descriptor: | PENTAETHYLENE GLYCOL, SULFATE ION, monoclonal anti-HIV-1 gp120 V3 antibody R20 heavy chain, ... | | Authors: | Pan, R.M, Kong, X.P. | | Deposit date: | 2013-03-16 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Rabbit Anti-HIV-1 Monoclonal Antibodies Raised by Immunization Can Mimic the Antigen-Binding Modes of Antibodies Derived from HIV-1-Infected Humans.
J.Virol., 87, 2013
|
|
4JO2
 
 | |
4JO1
 
 | |
