1A3C
 
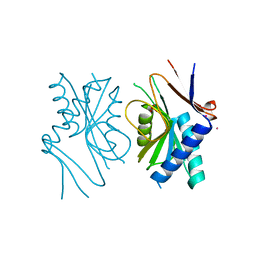 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, DIMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SAMARIUM (III) ION, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-01-20 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1A4X
 
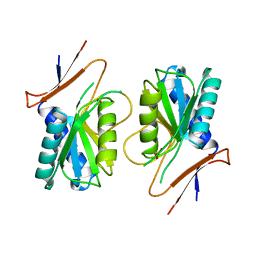 | | PYRR, THE BACILLUS SUBTILIS PYRIMIDINE BIOSYNTHETIC OPERON REPRESSOR, HEXAMERIC FORM | | Descriptor: | PYRIMIDINE OPERON REGULATORY PROTEIN PYRR, SULFATE ION | | Authors: | Tomchick, D.R, Turner, R.J, Switzer, R.W, Smith, J.L. | | Deposit date: | 1998-02-08 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
Structure, 6, 1998
|
|
1FGO
 
 | |
1FGM
 
 | |
1FGQ
 
 | |
1FGR
 
 | |
1FGT
 
 | |
1F8N
 
 | |
8DML
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt chenodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHENODEOXYCHOLIC ACID, ... | | Authors: | Tomchick, D.R, Orth, K, Zou, A.J. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Molecular determinants for differential activation of the bile acid receptor from the pathogen Vibrio parahaemolyticus.
J.Biol.Chem., 299, 2023
|
|
3L15
 
 | | Human Tead2 transcriptional factor | | Descriptor: | GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Tomchick, D.R, Luo, X, Tian, W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the YAP-binding domain of human TEAD2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3BEN
 
 | |
9B8D
 
 | | Structure of Legionella pneumophila Ceg10 | | Descriptor: | 1,2-ETHANEDIOL, Ceg10, PHOSPHATE ION | | Authors: | Tomchick, D.R, Heisler, D.B, Alto, N.M. | | Deposit date: | 2024-03-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exploiting bacterial effector proteins to uncover evolutionarily conserved antiviral host machinery.
Plos Pathog., 20, 2024
|
|
9B8E
 
 | | Structure of S-nitrosylated Legionella pneumophila Ceg10. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Heisler, D.B, Alto, N.M. | | Deposit date: | 2024-03-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploiting bacterial effector proteins to uncover evolutionarily conserved antiviral host machinery.
Plos Pathog., 20, 2024
|
|
6ANJ
 
 | | Synaptotagmin-7, C2A domain | | Descriptor: | ACETATE ION, CALCIUM ION, Synaptotagmin-7, ... | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6AO5
 
 | | Crystal structure of human MST2 in complex with SAV1 SARAH domain | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein salvador homolog 1, ... | | Authors: | Tomchick, D.R, Luo, X, Ni, L. | | Deposit date: | 2017-08-15 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK.
Elife, 6, 2017
|
|
6ANK
 
 | | Synaptotagmin-7, C2A- and C2B-domains | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6PWD
 
 | | Ewingella americana HopBF1 kinase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Type III effector HopBF1 | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Park, B.C. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A Bacterial Effector Mimics a Host HSP90 Client to Undermine Immunity.
Cell, 179, 2019
|
|
6PWG
 
 | | Ewingella americana HopBF1 kinase bound to AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Type III effector HopBF1 | | Authors: | Tomchick, D.R, Tagliabracci, V.S, Park, B.C. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Bacterial Effector Mimics a Host HSP90 Client to Undermine Immunity.
Cell, 179, 2019
|
|
4XDU
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein,a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, ADP bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-20 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
4XGX
 
 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase, Y60N mutant, ADP-inhibited | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XDR
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, D284A mutant, ADN bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
4XGW
 
 | | Crystal structure of Escherichia coli Flavin trafficking protein, an FMN transferase, E169K mutant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-02 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
5T8V
 
 | | Chaetomium thermophilum cohesin loader SCC2, C-terminal fragment | | Descriptor: | CITRIC ACID, Putative uncharacterized protein | | Authors: | Tomchick, D.R, Yu, H, Kikuchi, S, Ouyang, Z, Borek, D, Otwinowski, Z. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3KY9
 
 | | Autoinhibited Vav1 | | Descriptor: | Proto-oncogene vav, ZINC ION | | Authors: | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
6VVC
 
 | |
