1ZP0
 
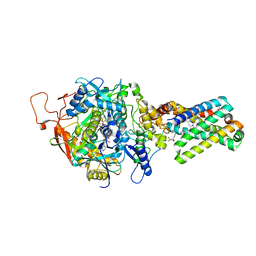 | | Crystal Structure of Mitochondrial Respiratory Complex II bound with 3-nitropropionate and 2-thenoyltrifluoroacetone | | Descriptor: | 3-NITROPROPANOIC ACID, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, FAD-binding protein, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-16 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZOY
 
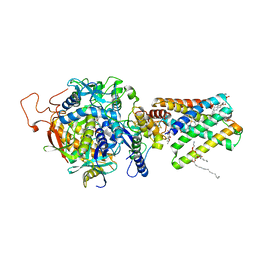 | | Crystal Structure of Mitochondrial Respiratory Complex II from porcine heart at 2.4 Angstroms | | Descriptor: | FAD-binding protein, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-15 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
4GXO
 
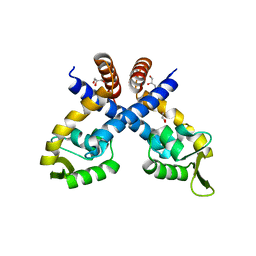 | | Crystal structure of Staphylococcus aureus protein SarZ mutant C13E | | Descriptor: | GLYCEROL, MarR family regulatory protein | | Authors: | Sun, F, Ding, Y, Ji, Q, Liang, Z, Deng, X, Wong, C.C, Yi, C, Zhang, L, Xie, S, Alvarez, S, Hicks, L.M, Luo, C, Jiang, H, Lan, L, He, C. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TPQ
 
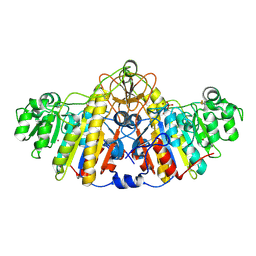 | | E. coli alkaline phosphatase D101A, D153A, R166S, E322A, K328A mutant | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Sunden, F, AlSadhan, I, Lyubimov, A.Y, Doukov, T, Swan, J, Herschlag, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
5X9C
 
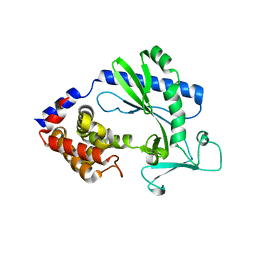 | |
5X9B
 
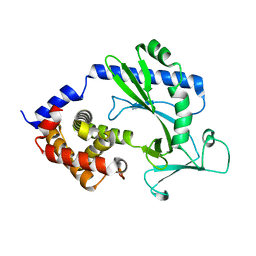 | |
5KCQ
 
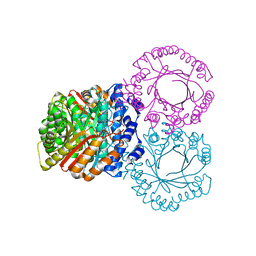 | |
5KDA
 
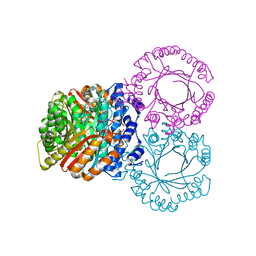 | |
5KCY
 
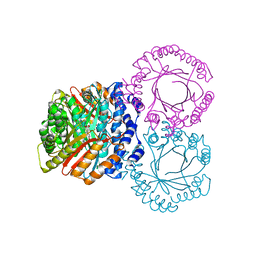 | |
5KCL
 
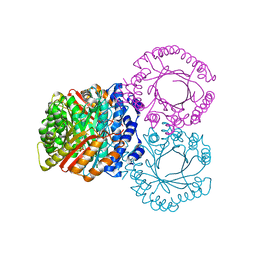 | |
5KD0
 
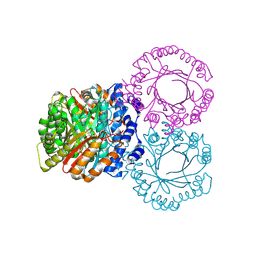 | |
5KD6
 
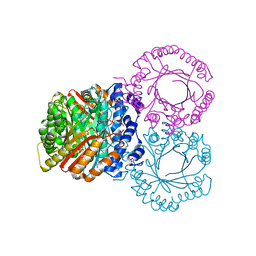 | |
5KCG
 
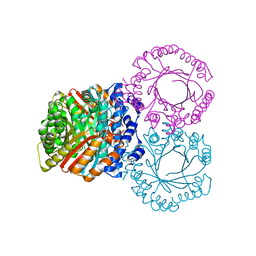 | |
7UPV
 
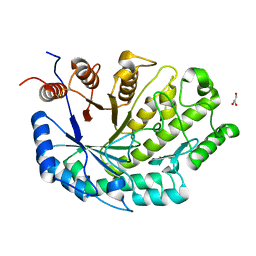 | |
4F1P
 
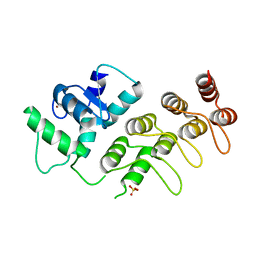 | | Crystal Structure of mutant S554D for ArfGAP and ANK repeat domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2012-05-07 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
3T9K
 
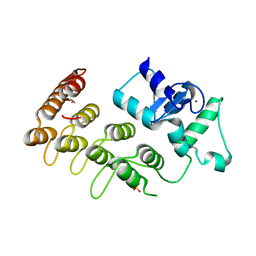 | | Crystal Structure of ACAP1 C-portion mutant S554D fused with integrin beta1 peptide | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1,Peptide from Integrin beta-1, SULFATE ION, ... | | Authors: | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
5TJ3
 
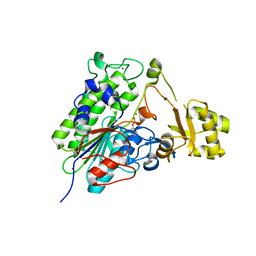 | | Crystal structure of wild type alkaline phosphatase PafA to 1.7A resolution | | Descriptor: | Alkaline phosphatase PafA, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, Ressl, S, Herschlag, D. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanistic and Evolutionary Insights from Comparative Enzymology of Phosphomonoesterases and Phosphodiesterases across the Alkaline Phosphatase Superfamily.
J.Am.Chem.Soc., 138, 2016
|
|
5TOO
 
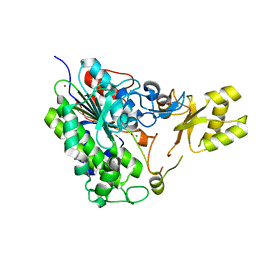 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | Descriptor: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
4Y9L
 
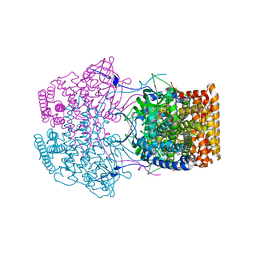 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | Authors: | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
3KH8
 
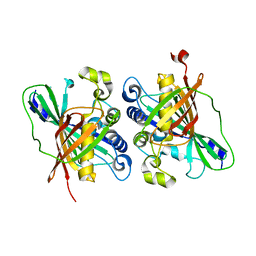 | | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici | | Descriptor: | MaoC-like dehydratase | | Authors: | Wang, H, Zhang, K, Guo, J, Zhou, Q, Zheng, X, Sun, F, Pang, H, Zhang, X. | | Deposit date: | 2009-10-30 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MaoC-like dehydratase from Phytophthora Capsici
To be Published
|
|
4UUQ
 
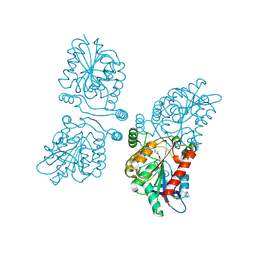 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
4Y9J
 
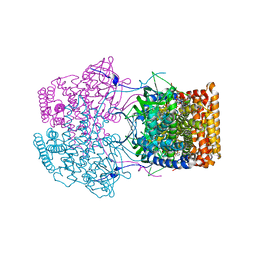 | |
6IUG
 
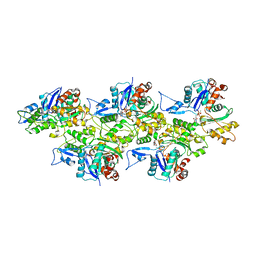 | | Cryo-EM structure of the plant actin filaments from Zea mays pollen | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, pollen F-actin | | Authors: | Ren, Z.H, Zhang, Y, Zhang, Y, He, Y.Q, Du, P.Z, Wang, Z.X, Sun, F, Ren, H.Y. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of Actin Filaments fromZea maysPollen.
Plant Cell, 31, 2019
|
|
3ZIF
 
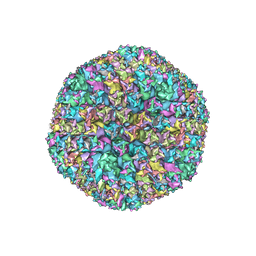 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
