4LF0
 
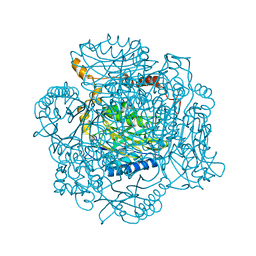 | | The E142D mutant of the amidase from Geobacillus pallidus | | Descriptor: | Aliphatic amidase | | Authors: | Sewell, B.T, Weber, B.W, Kimani, S.W, Cowan, D.A, Hunter, R, Venter, G.A, Gumbart, J.C, Thuku, R.N, Varsani, A. | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
4V4N
 
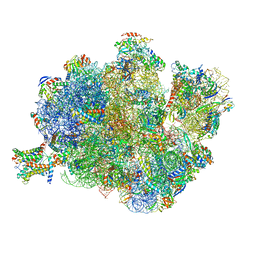 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3J45
 
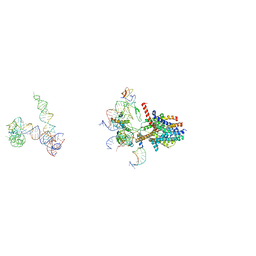 | | Structure of a non-translocating SecY protein channel with the 70S ribosome | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L23, 50S ribosomal protein L24, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3J46
 
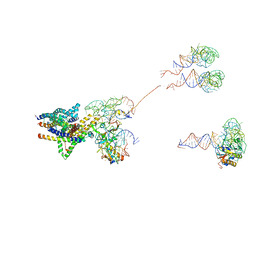 | | Structure of the SecY protein translocation channel in action | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L23P, ... | | Authors: | Akey, C.W, Park, E, Menetret, J.F, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-23 | | Last modified: | 2019-07-03 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
4V7I
 
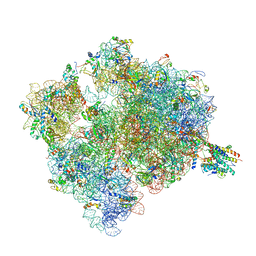 | | Ribosome-SecY complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gumbart, J.C, Trabuco, L.G, Schreiner, E, Villa, E, Schulten, K. | | Deposit date: | 2009-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Regulation of the protein-conducting channel by a bound ribosome.
Structure, 17, 2009
|
|
4X0K
 
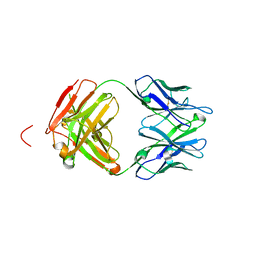 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain | | Authors: | Johnson, J.L, Lieberman, R.L. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YS0
 
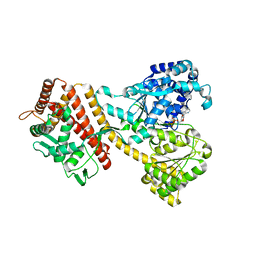 | |
4EYY
 
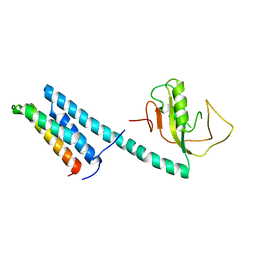 | | Crystal Structure of the IcmR-IcmQ complex from Legionella pneumophila | | Descriptor: | IcmQ, IcmR | | Authors: | Farelli, J.D, Gumbart, J, Akey, I.V, Amyot, W, Hempstead, A.D, Head, J.F, McKnight, C.J, Isberg, R.R, Akey, C.W. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | IcmQ in the Type 4b secretion system contains an NAD+ binding domain.
Structure, 21, 2013
|
|
5IXM
 
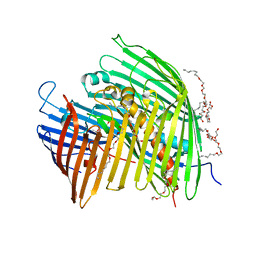 | | The LPS Transporter LptDE from Yersinia pestis, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Botos, I, Mayclin, S.J, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
5VR2
 
 | |
6WIL
 
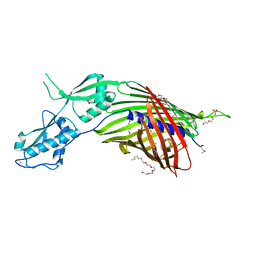 | | CdiB from Acinetobacter baumannii | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, DI(HYDROXYETHYL)ETHER, Hemolysin activator protein CdiB, ... | | Authors: | Guerin, J, Botos, I, Buchanan, S.K. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into toxin secretion by contact dependent growth inhibition transporters.
Elife, 9, 2020
|
|
6WIM
 
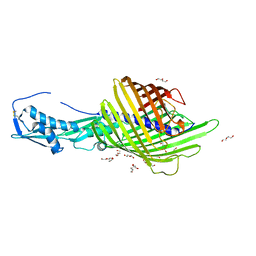 | | CdiB from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Outer membrane transporter CdiB | | Authors: | Guerin, J, Botos, I, Buchanan, S.K. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into toxin secretion by contact dependent growth inhibition transporters.
Elife, 9, 2020
|
|
8T62
 
 | |
8T63
 
 | |
8T61
 
 | |
8TXS
 
 | |
8E1M
 
 | |
8DOT
 
 | |
8FRR
 
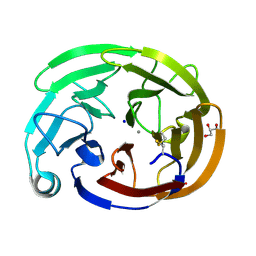 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
5IV8
 
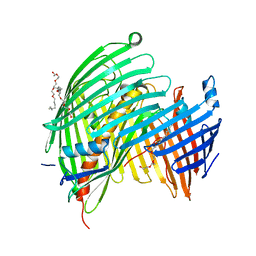 | | The LPS Transporter LptDE from Klebsiella pneumoniae, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS biosynthesis protein, LPS-assembly lipoprotein LptE | | Authors: | Botos, I, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.938 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
5IVA
 
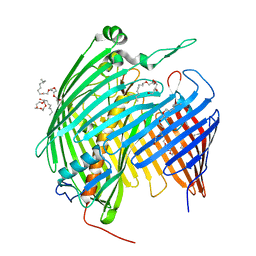 | |
5IV9
 
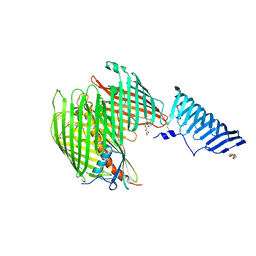 | | The LPS Transporter LptDE from Klebsiella pneumoniae, full-length | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD | | Authors: | Botos, I, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-20 | | Release date: | 2016-05-18 | | Last modified: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (4.369 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
6OU2
 
 | |
6OU3
 
 | |
6OU0
 
 | |
