4DD8
 
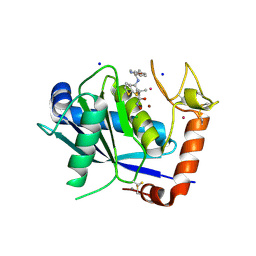 | | ADAM-8 metalloproteinase domain with bound batimastat | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2KR2
 
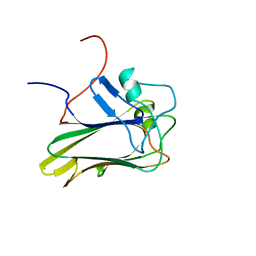 | |
8V44
 
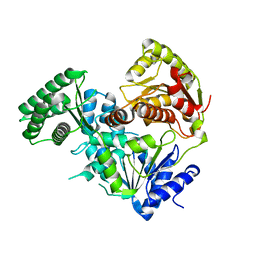 | |
2JWP
 
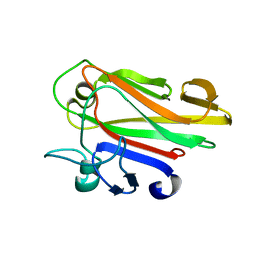 | | Malectin | | Descriptor: | Malectin | | Authors: | Schallus, T, Muhle-goll, C. | | Deposit date: | 2007-10-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
2K46
 
 | | Xenopus laevis malectin complexed with nigerose (Glcalpha1-3Glc) | | Descriptor: | MGC80075 protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Schallus, T. | | Deposit date: | 2008-05-28 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
2O10
 
 | |
2O13
 
 | |
6ZXU
 
 | |
6ZY1
 
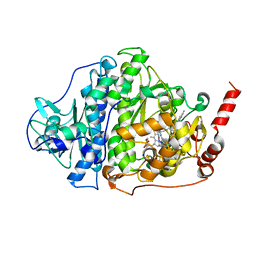 | |
6ZXX
 
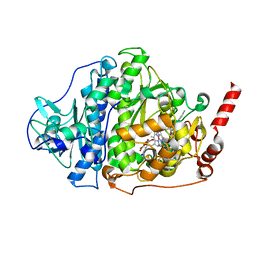 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged. | | Descriptor: | 3 bromo 4 hydroxybenzoic acid, 3,5-bis(bromanyl)-4-oxidanyl-benzoic acid, BROMIDE ION, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
6ZY0
 
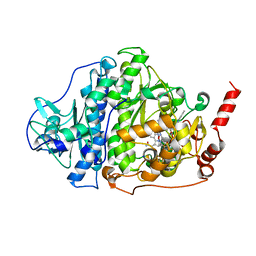 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged, K488Q variant | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
5E8N
 
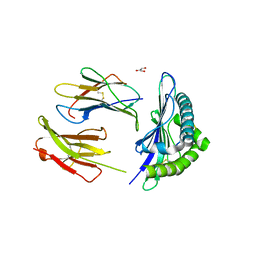 | | The structure of the TEIPP associated Trh4 peptide in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, GLYCEROL, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
5E8P
 
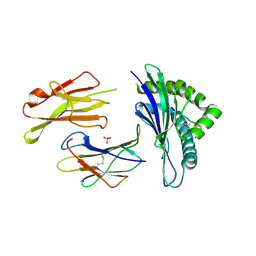 | | The structure of the TEIPP associated altered peptide ligand Trh4-p5NLE in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
5E8O
 
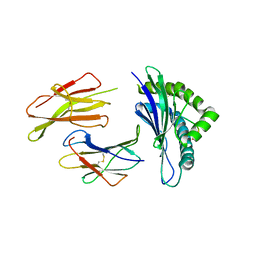 | | The structure of the TEIPP associated altered peptide ligand Trh4-p2ABU in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
2WLV
 
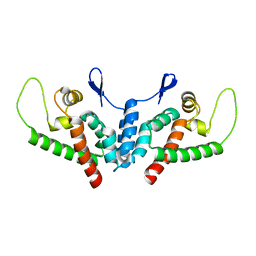 | | Structure of the N-terminal capsid domain of HIV-2 | | Descriptor: | GAG POLYPROTEIN | | Authors: | Price, A.J, Marzetta, F, Lammers, M, Ylinen, L.M.J, Schaller, T, Wilson, S.J, Towers, G.J, James, L.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Active Site Remodelling Switches HIV Specificity of Antiretroviral Trimcyp
Nat.Struct.Mol.Biol., 16, 2009
|
|
8RH1
 
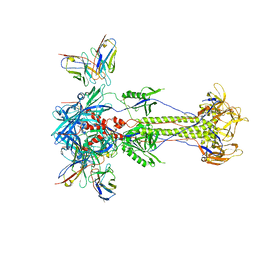 | | Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH2
 
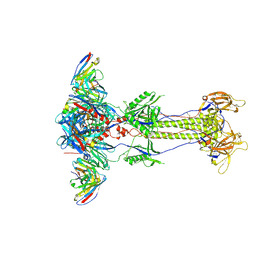 | | Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain, HDIT102 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RGZ
 
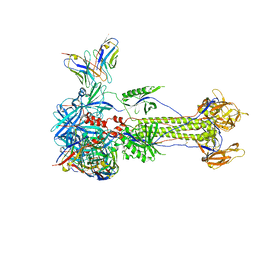 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH0
 
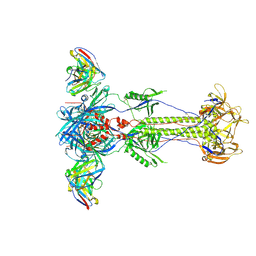 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
5A8G
 
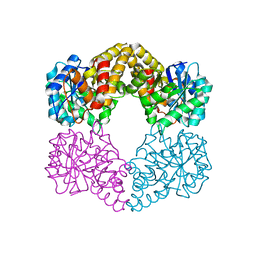 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
1EF9
 
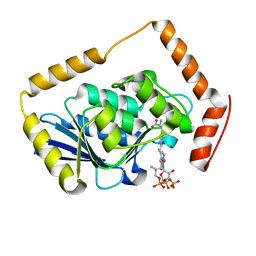 | | THE CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE COMPLEXED WITH 2S-CARBOXYPROPYL COA | | Descriptor: | 2-CARBOXYPROPYL-COENZYME A, METHYLMALONYL COA DECARBOXYLASE | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EF8
 
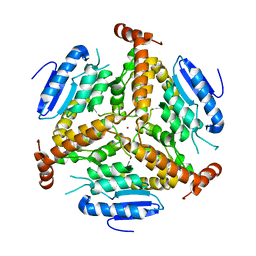 | | CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE | | Descriptor: | METHYLMALONYL COA DECARBOXYLASE, NICKEL (II) ION | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
2MA9
 
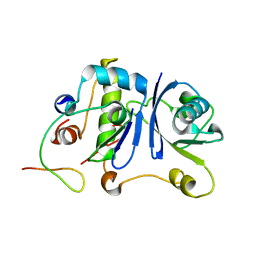 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
2WLW
 
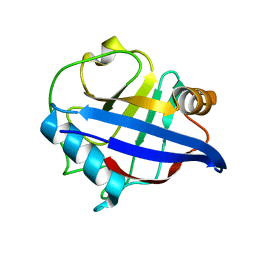 | | Structure of the N-terminal capsid domain of HIV-2 | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Price, A.J, Marzetta, F, Lammers, M, Ylinen, L.M.J, Schaller, T, Wilson, S.J, Towers, G.J, James, L.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active Site Remodelling Switches HIV Specificity of Antiretroviral Trimcyp
Nat.Struct.Mol.Biol., 16, 2009
|
|
2XR9
 
 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
