1HFK
 
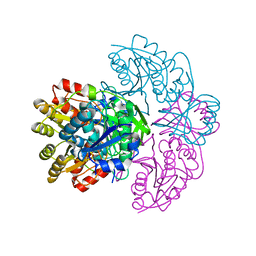 | | Asparaginase from Erwinia chrysanthemi, hexagonal form with weak sulfate | | Descriptor: | L-ASPARAGINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Lubkowski, J, Palm, G.J, Kozak, M, Jaskolski, M, Wlodawer, A. | | Deposit date: | 2000-12-05 | | Release date: | 2000-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structures of Two Highly Homologous Bacterial L-Asparaginases: A Case of Enantiomorphic Space Groups
Acta Crystallogr.,Sect.D, 57, 2001
|
|
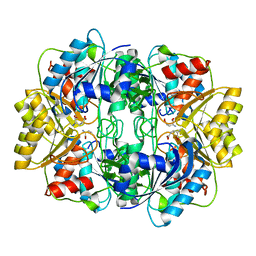
1HFW
 
 | |
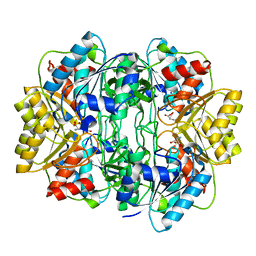
1HG1
 
 | |
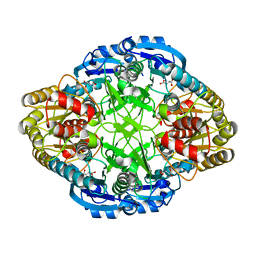
1HG0
 
 | |
5L04
 
 | |
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
6DF3
 
 | |
3PGA
 
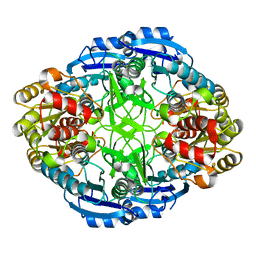 | | STRUCTURAL CHARACTERIZATION OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Ammon, H.L, Copeland, T.D, Swain, A.L. | | Deposit date: | 1994-07-19 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Pseudomonas 7A glutaminase-asparaginase.
Biochemistry, 33, 1994
|
|
2RA4
 
 | |
6PAE
 
 | |
6PAC
 
 | |
6PA3
 
 | |
6PA8
 
 | |
6PAB
 
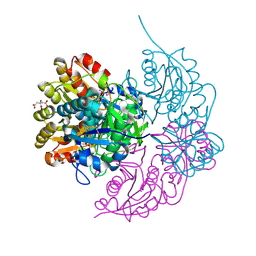 | |
6PA6
 
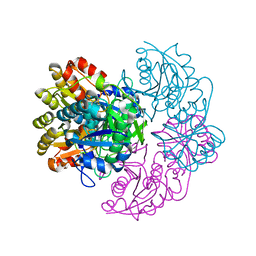 | |
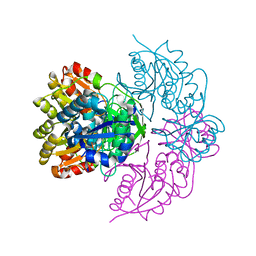
6PAA
 
 | |
5DUY
 
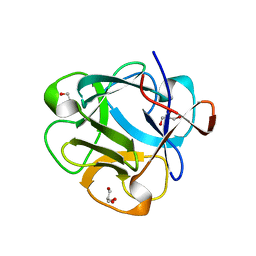 | | Structure of lectin from the sea mussel Crenomytilus grayanus | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Lubkowski, J, Jakob, M, O'Keefe, B, Wlodawer, A. | | Deposit date: | 2015-09-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a lectin from the sea mussel Crenomytilus grayanus (CGL).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1AGX
 
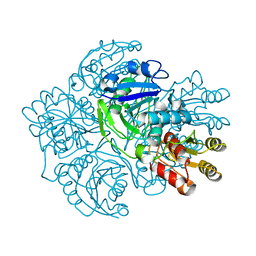 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
3HJ2
 
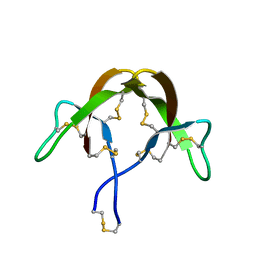 | |
3HJD
 
 | |
6PA5
 
 | |
6PA2
 
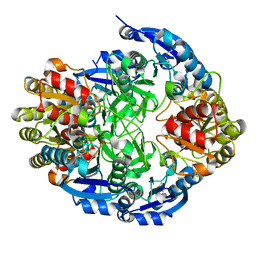 | |
6PA4
 
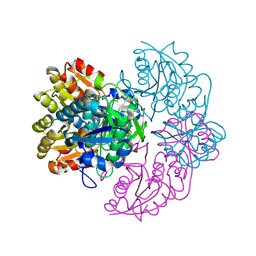 | |
3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | Authors: | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | Deposit date: | 2010-06-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1TOL
 
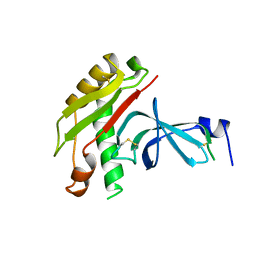 | | FUSION OF N-TERMINAL DOMAIN OF THE MINOR COAT PROTEIN FROM GENE III IN PHAGE M13, AND C-TERMINAL DOMAIN OF E. COLI PROTEIN-TOLA | | Descriptor: | PROTEIN (FUSION PROTEIN CONSISTING OF MINOR COAT PROTEIN, GLYCINE RICH LINKER, TOLA, ... | | Authors: | Lubkowski, J, Wlodawer, A, Hennecke, F, Plueckthun, A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA.
Structure Fold.Des., 7, 1999
|
|
