1KB9
 
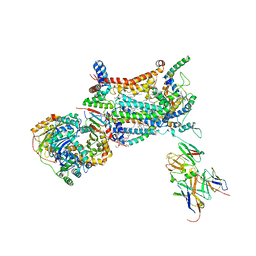 | | YEAST CYTOCHROME BC1 COMPLEX | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Lange, C, Nett, J.H, Trumpower, B.L, Hunte, C. | | Deposit date: | 2001-11-05 | | Release date: | 2002-09-18 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SPECIFIC ROLES OF PROTEIN-PHOSPHOLIPID INTERACTIONS IN THE YEAST CYTOCHROME BC1 COMPLEX STRUCTURE
EMBO J., 20, 2001
|
|
1KYO
 
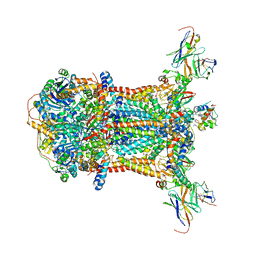 | |
1TKW
 
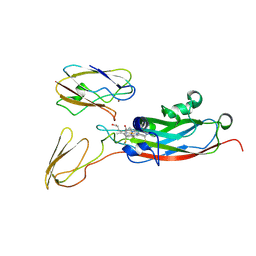 | | The transient complex of poplar plastocyanin with turnip cytochrome f determined with paramagnetic NMR | | Descriptor: | COPPER (II) ION, Cytochrome f, HEME C, ... | | Authors: | Lange, C, Cornvik, T, Diaz-Moreno, I, Ubbink, M. | | Deposit date: | 2004-06-09 | | Release date: | 2005-05-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The transient complex of poplar plastocyanin with cytochrome f: effects of ionic strength and pH
Biochim.Biophys.Acta, 1707, 2005
|
|
3JZ4
 
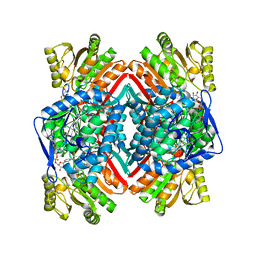 | | Crystal structure of E. coli NADP dependent enzyme | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | Authors: | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2009-09-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
8TJQ
 
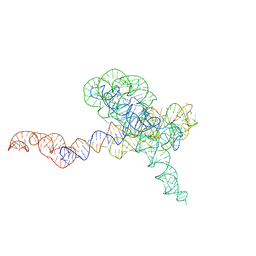 | |
8TJU
 
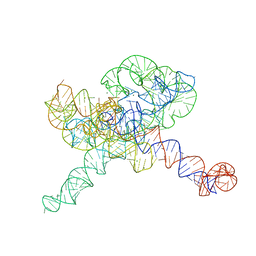 | | Tetrahymena Ribozyme scaffolded TABV xrRNA | | Descriptor: | DNA/RNA (416-MER), MAGNESIUM ION | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8TJV
 
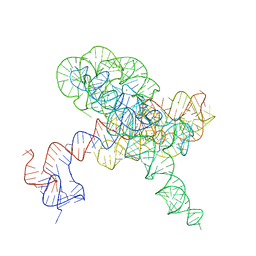 | |
8TJX
 
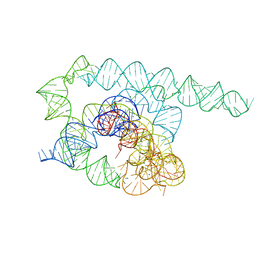 | | Tetrahymena Ribozyme cryo-EM scaffold | | Descriptor: | MAGNESIUM ION, RNA (440-MER) | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
3VP6
 
 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | Descriptor: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | Authors: | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
4ZHX
 
 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
5EZV
 
 | |
8GBD
 
 | | Homo sapiens Zalpha mutant - P193A | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Vogeli, B, Nichols, P.J, Henen, M, Vicens, Q. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8GBC
 
 | | Homo sapiens Zalpha mutant - N173S | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Nichols, P.J, Henen, M, Vicens, Q, Vogeli, B. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
2JVF
 
 | | Solution structure of M7, a computationally-designed artificial protein | | Descriptor: | de novo protein M7 | | Authors: | Stordeur, C, Dalluege, R, Birkenmeier, O, Wienk, H, Rudolph, R, Lange, C, Luecke, C. | | Deposit date: | 2007-09-19 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the artificial protein M7 matches the computationally designed model
Proteins, 72, 2008
|
|
2YNM
 
 | | Structure of the ADPxAlF3-Stabilized Transition State of the Nitrogenase-like Dark-Operative Protochlorophyllide Oxidoreductase Complex from Prochlorococcus marinus with Its Substrate Protochlorophyllide a | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Krausze, J, Lange, C, Heinz, D.W, Moser, J. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1EZV
 
 | | STRUCTURE OF THE YEAST CYTOCHROME BC1 COMPLEX CO-CRYSTALLIZED WITH AN ANTIBODY FV-FRAGMENT | | Descriptor: | 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CYTOCHROME B, CYTOCHROME C1, ... | | Authors: | Hunte, C, Koepke, J, Lange, C, Rossmanith, T, Michel, H. | | Deposit date: | 2000-05-12 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment.
Structure Fold.Des., 8, 2000
|
|
2V5Q
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE PLK-1 KINASE DOMAIN IN COMPLEX WITH A SELECTIVE DARPIN | | Descriptor: | DESIGN ANKYRIN REPEAT PROTEIN, SERINE/THREONINE-PROTEIN KINASE PLK1 | | Authors: | Bandeiras, T.M, Hillig, R.C, Matias, P.M, Eberspaecher, U, Fanghaenel, J, Thomaz, M, Miranda, S, Crusius, K, Puetter, V, Amstutz, P, Gulotti-Georgieva, M, Binz, H.K, Holz, C, Schmitz, A.A.P, Lang, C, Donner, P, Egner, U, Carrondo, M.A, Mueller-Tiemann, B. | | Deposit date: | 2007-07-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of wild-type Plk-1 kinase domain in complex with a selective DARPin.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
1SZ7
 
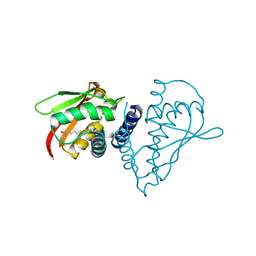 | | Crystal structure of Human Bet3 | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3 | | Authors: | Turnbull, A.P, Prinz, B, Holz, C, Behlke, J, Schultchen, J, Delbrueck, H, Niesen, F.H, Lang, C, Heinemann, U. | | Deposit date: | 2004-04-05 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization
Embo J., 24, 2005
|
|
7B4R
 
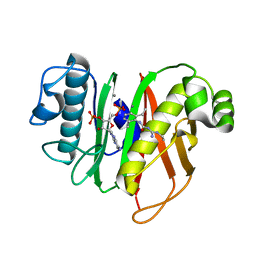 | | Structure of the 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus in complex with Coenzyme A and N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Maveyraud, L, Carivenc, C, Blanger, C, Mourey, L. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7MYJ
 
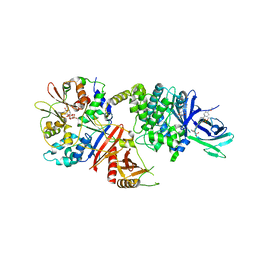 | | Structure of full length human AMPK (a2b1g1) in complex with a small molecule activator MSG011 | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Ovens, A.J, Gee, Y.S, Ling, N.X.Y, Waters, N.J, Yu, D, Scott, J.W, Parker, M.W, Hoffman, N.J, Kemp, B.E, Baell, J.B, Oakhill, J.S, Langendorf, C.G. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Biochem.J., 479, 2022
|
|
6B1U
 
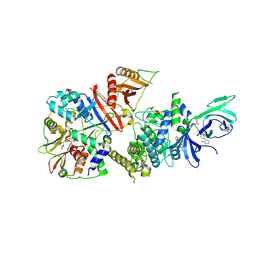 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6B2E
 
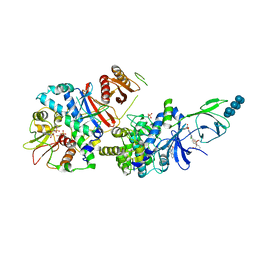 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6EF5
 
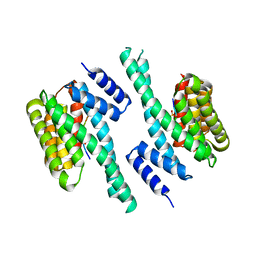 | | 14-3-3 with peptide | | Descriptor: | 14-3-3 protein zeta/delta, ARG-SER-LEU-SEP-ALA-PRO-GLY, LYS-LEU-SEP-LEU-GLN | | Authors: | Dhagat, U, Langendorf, C.G, Parker, M.W.P, Oakhill, J.S, Scott, J.W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | 14-3-3 with peptide
To Be Published
|
|
2R9Y
 
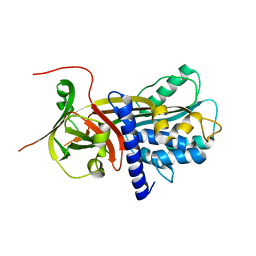 | | Structure of antiplasmin | | Descriptor: | Alpha-2-antiplasmin | | Authors: | Law, R.H.P, Sofian, T, Kan, W.T, Horvath, A.J, Hitchen, C.R, Langendorf, C.G, Buckle, A.M, Whisstock, J.C, Coughlin, P.B. | | Deposit date: | 2007-09-14 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray crystal structure of the fibrinolysis inhibitor {alpha}2-antiplasmin
Blood, 111, 2008
|
|
6BX6
 
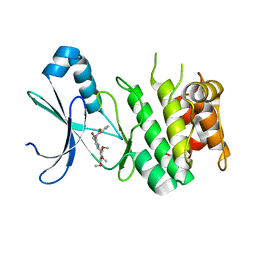 | | AMP-Activated protein kinase (AMPK) inhibition by SBI-0206965: alpha 2 kinase domain bound to SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Dite, T.A, Langendorf, C.G, Scott, J.W, Oakhill, J.S. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965.
J. Biol. Chem., 293, 2018
|
|
