2GTV
 
 | |
2LUM
 
 | |
8GBD
 
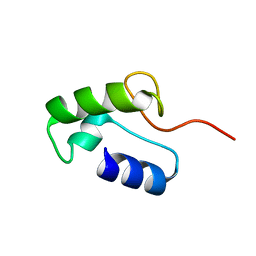 | | Homo sapiens Zalpha mutant - P193A | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Vogeli, B, Nichols, P.J, Henen, M, Vicens, Q. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
8GBC
 
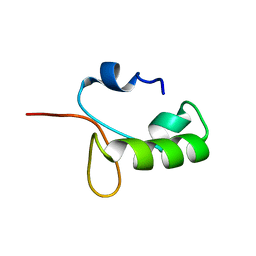 | | Homo sapiens Zalpha mutant - N173S | | Descriptor: | Double-stranded RNA-specific adenosine deaminase | | Authors: | Langeberg, C.J, Nichols, P.J, Henen, M, Vicens, Q, Vogeli, B. | | Deposit date: | 2023-02-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Differential Structural Features of Two Mutant ADAR1p150 Z alpha Domains Associated with Aicardi-Goutieres Syndrome.
J.Mol.Biol., 435, 2023
|
|
7SUQ
 
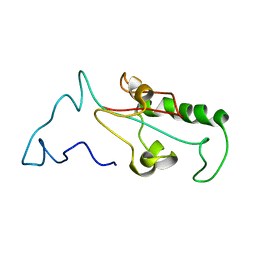 | |
7SUR
 
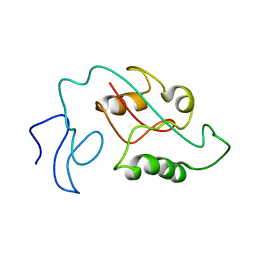 | |
7SA5
 
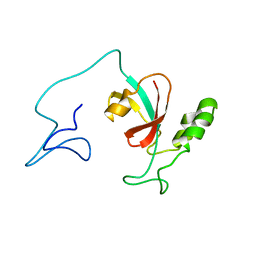 | | Two-state solution NMR structure of Apo Pin1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Born, A, Vogeli, B. | | Deposit date: | 2021-09-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reconstruction of Coupled Intra- and Interdomain Protein Motion from Nuclear and Electron Magnetic Resonance.
J.Am.Chem.Soc., 143, 2021
|
|
6BY5
 
 | | Two-State 14-mer UUCG Tetraloop calculated from Exact NOEs (State one: Conformers 1-5, State Two: Conformers 6-10) | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
6BY4
 
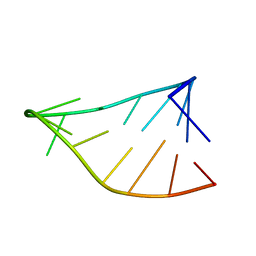 | | Single-State 14-mer UUCG Tetraloop calculated from Exact NOEs | | Descriptor: | RNA (5'-R(P*GP*GP*CP*AP*CP*UP*UP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Nichols, P.J, Henen, M.A, Born, A, Strotz, D, Guntert, P, Vogeli, B. | | Deposit date: | 2017-12-19 | | Release date: | 2018-06-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | High-resolution small RNA structures from exact nuclear Overhauser enhancement measurements without additional restraints.
Commun Biol, 1, 2018
|
|
6SVE
 
 | | Protein allostery of the WW domain at atomic resolution: pCdc25C bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SVH
 
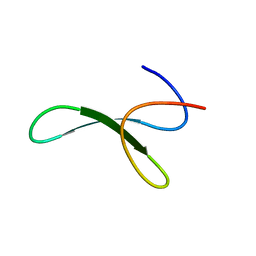 | | Protein allostery of the WW domain at atomic resolution: FFpSPR bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
