7TTW
 
 | | 50S ribosomal subunit from Staphylococcus aureus containing double mutation in uL3 imparting linezolid resistance | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Piper, S, Johnson, R. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A Structurally Characterized Staphylococcus aureus Evolutionary Escape Route from Treatment with the Antibiotic Linezolid.
Microbiol Spectr, 10, 2022
|
|
6YSN
 
 | | Human TRPC5 in complex with Pico145 (HC-608) | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 5 | | Authors: | Wright, D.J, Johnson, R.M, Muench, S.P, Bon, R.S. | | Deposit date: | 2020-04-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site.
Commun Biol, 3, 2020
|
|
6Z12
 
 | | Salmonella AcrB solubilised in the SMA copolymer | | Descriptor: | Efflux pump membrane transporter | | Authors: | Muench, s.p, Johnson, R.M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure and Molecular Dynamics Analysis of the Fluoroquinolone Resistant Mutant of the AcrB Transporter fromSalmonella.
Microorganisms, 8, 2020
|
|
5L2X
 
 | | Crystal structure of human PrimPol ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*GP*TP*AP*GP*CP*(DDG))-3'), ... | | Authors: | Rechkoblit, O, Gupta, Y.K, Malik, R, Rajashankar, K.R, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of human PrimPol, a DNA polymerase with primase activity.
Sci Adv, 2, 2016
|
|
3PZP
 
 | | Human DNA polymerase kappa extending opposite a cis-syn thymine dimer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*CP*A)-3', 5'-D(*TP*TP*CP*CP*(TTD)P*GP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Vasquez-Del Carpio, R, Silverstein, T.D, Lone, S, Johnson, R.E, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.336 Å) | | Cite: | Role of human DNA polymerase kappa in extension opposite from a cis-syn thymine dimer.
J.Mol.Biol., 408, 2011
|
|
1HCR
 
 | |
1IJW
 
 | | Testing the Water-Mediated Hin Recombinase DNA Recognition by Systematic Mutations. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*(CBR)P*TP*TP*AP*TP*CP*AP*AP*AP*AP*AP*C)-3', 5'-D(*TP*GP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*GP*A)-3', ... | | Authors: | Chiu, T.K, Sohn, C, Johnson, R.C, Dickerson, R.E. | | Deposit date: | 2001-04-30 | | Release date: | 2002-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Testing water-mediated DNA recognition by the Hin recombinase.
EMBO J., 21, 2002
|
|
3TQ1
 
 | | Human DNA Polymerase eta in binary complex with DNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*TP*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Ummat, A, Silverstein, T.D, Jain, R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2011-09-08 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Human DNA Polymerase Eta Is Pre-Aligned for dNTP Binding and Catalysis.
J.Mol.Biol., 415, 2012
|
|
6QQ6
 
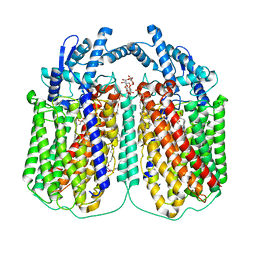 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
7TYF
 
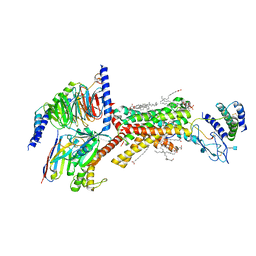 | | Human Amylin1 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TZF
 
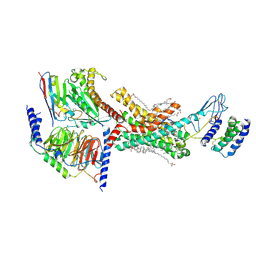 | | Human Amylin3 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYY
 
 | | Human Amylin2 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYX
 
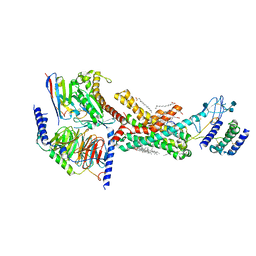 | | Human Amylin2 Receptor in complex with Gs and rat amylin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYN
 
 | | Calcitonin Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYI
 
 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, CT-like state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
4IHW
 
 | |
4IHY
 
 | |
4IHX
 
 | |
4IHV
 
 | |
7TYW
 
 | | Human Amylin1 Receptor in complex with Gs and salmon calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin receptor, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYL
 
 | | Calcitonin Receptor in complex with Gs and rat amylin peptide, bypass motif | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYH
 
 | | Human Amylin2 Receptor in complex with Gs and human calcitonin peptide | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-13 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
7TYO
 
 | | Calcitonin receptor in complex with Gs and human calcitonin peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Calcitonin, ... | | Authors: | Cao, J, Belousoff, M.J, Johnson, R.M, Wootten, D.L, Sexton, P.M. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-23 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural basis for amylin receptor phenotype.
Science, 375, 2022
|
|
3ITE
 
 | | The third adenylation domain of the fungal SidN non-ribosomal peptide synthetase | | Descriptor: | CHLORIDE ION, SULFATE ION, SidN siderophore synthetase | | Authors: | Lee, T.V, Lott, J.S, Johnson, R.D, Johnson, L.J, Arcus, V.L. | | Deposit date: | 2009-08-28 | | Release date: | 2009-11-17 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis
J.Biol.Chem., 285, 2010
|
|
7LCI
 
 | | PF 06882961 bound to the glucagon-like peptide-1 receptor (GLP-1R):Gs complex | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Johnson, R.M, Drulyte, I, Yu, L, Kotecha, A, Danev, R, Wootten, D, Zhang, X, Sexton, P.M. | | Deposit date: | 2021-01-11 | | Release date: | 2021-01-20 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Evolving cryo-EM structural approaches for GPCR drug discovery.
Structure, 29, 2021
|
|
