6CJ1
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JWG071 | | Descriptor: | 1,2-ETHANEDIOL, 11-[(2R)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, 11-[(2S)-butan-2-yl]-2-({2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidine-1-carbonyl]phenyl}amino)-5-methyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, ... | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
8G2Y
 
 | | Cryo-EM structure of ADGRF1 coupled to miniGs/q | | Descriptor: | Adhesion G-protein-coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jones, D, Rawson, S, Blacklow, S. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Tethered agonist activated ADGRF1 structure and signalling analysis reveal basis for G protein coupling.
Nat Commun, 14, 2023
|
|
4WIV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a novel inhibitor UMB32 (N-TERT-BUTYL-2-[4-(3,5-DIMETHYL-1,2-OXAZOL-4-YL) PHENYL]IMIDAZO[1,2-A]PYRAZIN-3-AMINE) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, X, Blacklow, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biased multicomponent reactions to develop novel bromodomain inhibitors.
J.Med.Chem., 57, 2014
|
|
3V79
 
 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
6CRF
 
 | | Crystal Structure of Shp2 E76K GOF Mutant in the Open Conformation | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2018-03-17 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition.
Nat Commun, 9, 2018
|
|
6U9S
 
 | | Crystal structure of human CD81 large extracellular loop in complex with 5A6 Fab | | Descriptor: | 5A6 FAB Heavy Chain, 5A6 FAB Light Chain, CD81 antigen, ... | | Authors: | Susa, K.J, Seegar, T.C.M, Blacklow, S.C.B, Kruse, A.C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking.
Elife, 9, 2020
|
|
7RCT
 
 | |
6UFY
 
 | | B. theta Bile Salt Hydrolase | | Descriptor: | Choloylglycine hydrolase | | Authors: | Seegar, T.C.M. | | Deposit date: | 2019-09-25 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Development of a covalent inhibitor of gut bacterial bile salt hydrolases.
Nat.Chem.Biol., 16, 2020
|
|
6UH4
 
 | | B. theta Bile Salt Hydrolase with covalent inhibitor | | Descriptor: | (5R,6R)-6-[(1S,2R,4aS,4bS,7R,8aS,10R,10aS)-7,10-dihydroxy-1,2,4b-trimethyltetradecahydrophenanthren-2-yl]-5-methylheptan-2-one, Choloylglycine hydrolase | | Authors: | Seegar, T.C.M. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Development of a covalent inhibitor of gut bacterial bile salt hydrolases.
Nat.Chem.Biol., 16, 2020
|
|
5CZX
 
 | | Crystal structure of Notch3 NRR in complex with 20358 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 20358 Fab heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
5CZV
 
 | | Crystal structure of Notch3 NRR in complex with 20350 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 20350 heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
2FTU
 
 | | solution structure of domain 3 of RAP | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein, domain 3 | | Authors: | Lee, D, Walsh, J.D, Wang, Y.-X. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-09 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi.
Mol.Cell, 22, 2006
|
|
1TPW
 
 | | TRIOSEPHOSPHATE ISOMERASE DRINKS WATER TO KEEP HEALTHY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Stock, A.M, Narayana, N, Xuong, Ng.H, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-11-07 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of water in the catalytic efficiency of triosephosphate isomerase.
Biochemistry, 38, 1999
|
|
1SZT
 
 | |
6BDZ
 
 | | ADAM10 Extracellular Domain Bound by the 11G2 Fab | | Descriptor: | 11G2 Fab Heavy Chain, 11G2 Fab Light Chain, CALCIUM ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BE6
 
 | | ADAM10 Extracellular Domain | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 10, SULFATE ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BMV
 
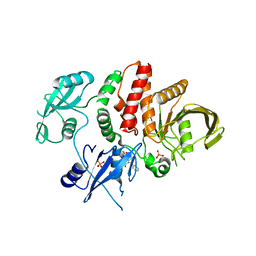 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP504 | | Descriptor: | 3-{4-[(2-chlorophenyl)methyl]-5-oxo-4,5-dihydro[1,2,4]triazolo[4,3-a]quinazolin-1-yl}-4-hydroxybenzoic acid, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMU
 
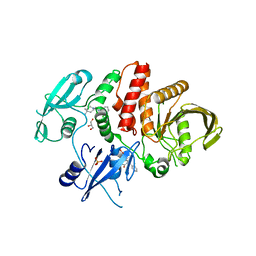 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMY
 
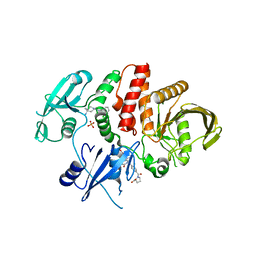 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP844 | | Descriptor: | 1-(3-chloro-4-{[1-(2-hydroxy-3-methoxyphenyl)-5-oxo[1,2,4]triazolo[4,3-a]quinazolin-4(5H)-yl]methyl}benzene-1-carbonyl)-L-proline, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMR
 
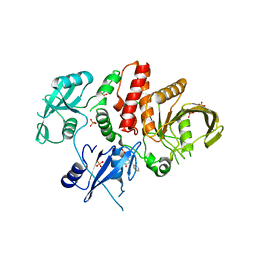 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMX
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP844 | | Descriptor: | 1-(3-chloro-4-{[1-(2-hydroxy-3-methoxyphenyl)-5-oxo[1,2,4]triazolo[4,3-a]quinazolin-4(5H)-yl]methyl}benzene-1-carbonyl)-L-proline, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMW
 
 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP504 | | Descriptor: | 3-{4-[(2-chlorophenyl)methyl]-5-oxo-4,5-dihydro[1,2,4]triazolo[4,3-a]quinazolin-1-yl}-4-hydroxybenzoic acid, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6CRG
 
 | | Crystal Structure of Shp2 E76K GOF Mutant in complex with SHP099 | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2018-03-17 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural reorganization of SHP2 by oncogenic mutations and implications for oncoprotein resistance to allosteric inhibition.
Nat Commun, 9, 2018
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
