1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
5JQW
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-05 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
5KBP
 
 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | Descriptor: | Glycosyl hydrolase, family 38, SULFATE ION | | Authors: | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
1LSL
 
 | | Crystal Structure of the Thrombospondin-1 Type 1 Repeats | | Descriptor: | Thrombospondin 1, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Tan, K, Duquette, M, Liu, J, Dong, Y, Zhang, R, Joachimiak, A, Lawler, J, Wang, J.-H. | | Deposit date: | 2002-05-17 | | Release date: | 2002-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the TSP-1 type 1 repeats: a novel
layered fold and its biological implication.
J.Cell Biol., 159, 2002
|
|
1SZT
 
 | |
2ES3
 
 | |
4YYF
 
 | | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155 | | Descriptor: | ACETATE ION, Beta-N-acetylhexosaminidase, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a glycosyl hydrolase of GH3 family member from [Mycobacterium smegmatis str. MC2 155
To Be Published
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
6PXA
 
 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | Descriptor: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | Authors: | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid
To Be Published
|
|
2LM9
 
 | |
8TFG
 
 | |
1Z78
 
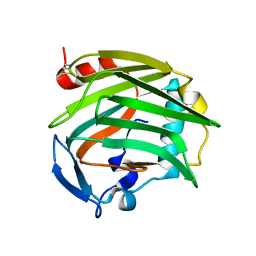 | |
1ZA4
 
 | |
7N6H
 
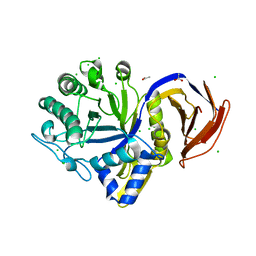 | |
7N6O
 
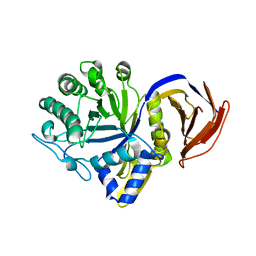 | |
6WHL
 
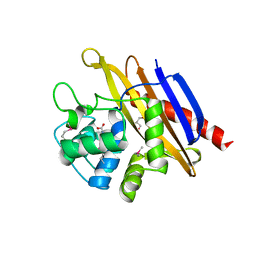 | |
2ERF
 
 | |
6XOA
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation
To Be Published
|
|
4R7Q
 
 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
6E4B
 
 | |
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
2HKU
 
 | | Structural Genomics, the crystal structure of a putative transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, A putative transcriptional regulator, TETRAETHYLENE GLYCOL | | Authors: | Tan, K, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-05 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
2I5E
 
 | | Crystal Structure of a Protein of Unknown Function MM2497 from Methanosarcina mazei Go1, Probable Nucleotidyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Hypothetical protein MM_2497 | | Authors: | Tan, K, Du, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a hypothetical protein MM_2497 from Methanosarcina mazei Go1
To be Published
|
|
2IA9
 
 | | Structural Genomics, the crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative septation protein spoVG, SULFATE ION | | Authors: | Tan, K, Borovilos, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-07 | | Release date: | 2006-10-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of SpoVG from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
