5OH0
 
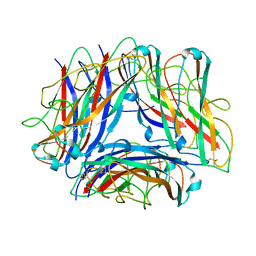 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
1QSY
 
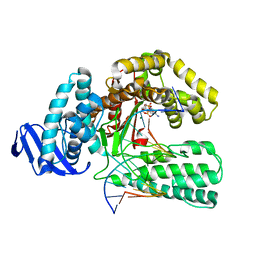 | | DDATP-Trapped closed ternary complex of the large fragment of DNA Polymerase I from thermus aquaticus | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, 5'-D(*AP*TP*TP*GP*CP*GP*CP*CP*TP*P*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DA))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1NZV
 
 | |
1QTM
 
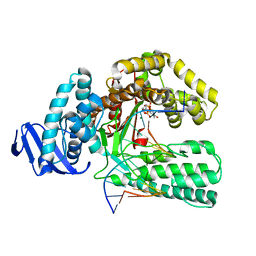 | | DDTTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(2DT))-3', DNA POLYMERASE I, ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-28 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1PDK
 
 | | PAPD-PAPK CHAPERONE-PILUS SUBUNIT COMPLEX FROM E.COLI P PILUS | | Descriptor: | PROTEIN (CHAPERONE PROTEIN PAPD), PROTEIN (PROTEIN PAPK) | | Authors: | Fuetterer, K, Sauer, F.G, Hultgren, S.J, Waksman, G. | | Deposit date: | 1999-03-29 | | Release date: | 1999-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of chaperone function and pilus biogenesis.
Science, 285, 1999
|
|
1IIC
 
 | |
1KC2
 
 | |
1IID
 
 | | Crystal Structure of Saccharomyces cerevisiae N-myristoyltransferase with Bound S-(2-oxo)pentadecylCoA and the Octapeptide GLYASKLA | | Descriptor: | NICKEL (II) ION, Octapeptide GLYASKLA, Peptide N-myristoyltransferase, ... | | Authors: | Farazi, T.A, Gordon, J.I, Waksman, G. | | Deposit date: | 2001-04-22 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis.
Biochemistry, 40, 2001
|
|
1KAW
 
 | | STRUCTURE OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) | | Descriptor: | SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Raghunathan, S, Waksman, G. | | Deposit date: | 1996-12-06 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1MH0
 
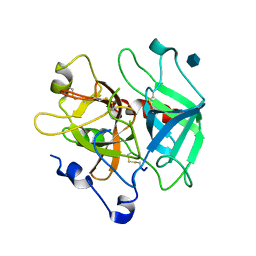 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pineda, A.O, Savvides, S, Waksman, G, Di Cera, E. | | Deposit date: | 2002-08-18 | | Release date: | 2002-11-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the anticoagulant slow form of thrombin
J.Biol.Chem., 277, 2002
|
|
1KTQ
 
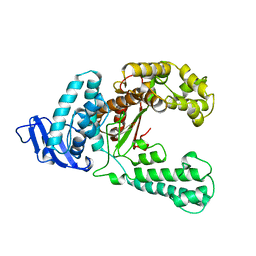 | | DNA POLYMERASE | | Descriptor: | DNA POLYMERASE I | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1995-08-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1TQ0
 
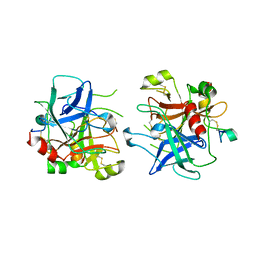 | | Crystal structure of the potent anticoagulant thrombin mutant W215A/E217A in free form | | Descriptor: | Prothrombin | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1TQ7
 
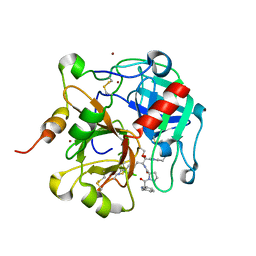 | | Crystal structure of the anticoagulant thrombin mutant W215A/E217A bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.-W, Caccia, S, Savvides, S.N, Waksman, G, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The anticoagulant thrombin mutant W215A/E217A has a collapsed primary specificity pocket
J.Biol.Chem., 279, 2004
|
|
1UAA
 
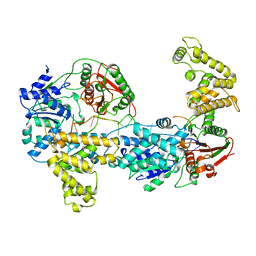 | | E. COLI REP HELICASE/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), PROTEIN (ATP-DEPENDENT DNA HELICASE REP.) | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.
Cell(Cambridge,Mass.), 90, 1997
|
|
1S7M
 
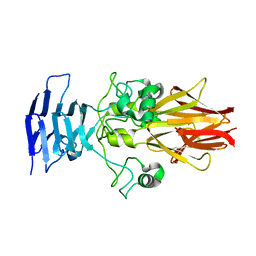 | | Crystal Structure of HiaBD1 | | Descriptor: | Hia | | Authors: | Yeo, H.J, Cotter, S.E, Laarmann, S, Juehne, T, St Geme, J.W, Waksman, G. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter.
Embo J., 23, 2004
|
|
1R8I
 
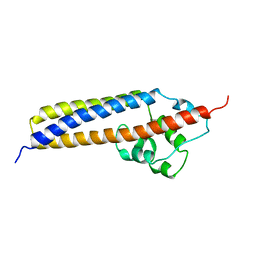 | | Crystal structure of TraC | | Descriptor: | TraC | | Authors: | Yeo, H.-J, Yuan, Q, Beck, M.R, Baron, C, Waksman, G. | | Deposit date: | 2003-10-24 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1SRU
 
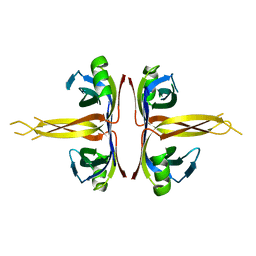 | | Crystal structure of full length E. coli SSB protein | | Descriptor: | Single-strand binding protein | | Authors: | Savvides, S.N, Raghunathan, S, Fuetterer, K, Kozlov, A.G, Lohman, T.M, Waksman, G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA.
Protein Sci., 13, 2004
|
|
1THP
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225P MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B7X
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225I MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | PROTEIN (INHIBITOR), PROTEIN (THROMBIN HEAVY CHAIN), PROTEIN (THROMBIN LIGHT CHAIN) | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-25 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1A81
 
 | |
1EKB
 
 | | THE SERINE PROTEASE DOMAIN OF ENTEROPEPTIDASE BOUND TO INHIBITOR VAL-ASP-ASP-ASP-ASP-LYS-CHLOROMETHANE | | Descriptor: | ENTEROPEPTIDASE, VAL-ASP-ASP-ASP-ASP-LYK PEPTIDE, ZINC ION | | Authors: | Fuetterer, K, Lu, D, Sadler, J.E, Waksman, G. | | Deposit date: | 1999-05-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enteropeptidase light chain complexed with an analog of the trypsinogen activation peptide.
J.Mol.Biol., 292, 1999
|
|
1EYG
 
 | |
3L48
 
 | |
2WMP
 
 | | Structure of the E. coli chaperone PapD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | CHAPERONE PROTEIN PAPD, PAPG PROTEIN | | Authors: | Ford, B.A, Verger, D, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2009-07-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Papd-Papgii Pilin Complex Reveals an Open and Flexible P5 Pocket.
J.Bacteriol., 194, 2012
|
|
1F6M
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
