4DST
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSO
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1KMX
 
 | |
1LB7
 
 | | IGF-F1-1, A PEPTIDE ANTAGONIST OF IGF-1 | | Descriptor: | IGF-1 ANTAGONIST F1-1 | | Authors: | Deshayes, K, Schaffer, M.L, Skelton, N.J, Nakamura, G.R, Kadkhodayan, S, Sidhu, S.S. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Rapid identification of small binding motifs with high-throughput phage display: discovery of peptidic antagonists of IGF-1 function.
Chem.Biol., 9, 2002
|
|
1IMX
 
 | | 1.8 Angstrom crystal structure of IGF-1 | | Descriptor: | BROMIDE ION, Insulin-like Growth Factor 1A, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE | | Authors: | Vajdos, F.F, Ultsch, M, Schaffer, M.L, Deshayes, K.D, Liu, J, Skelton, N.J, de Vos, A.M. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions.
Biochemistry, 40, 2001
|
|
1EDJ
 
 | |
1EDK
 
 | |
1EDL
 
 | |
1EDI
 
 | |
4NMX
 
 | |
3OCB
 
 | | Akt1 kinase domain with pyrrolopyrimidine inhibitor | | Descriptor: | (2S)-2-(4-chlorobenzyl)-3-oxo-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperazin-1-yl]propan-1-amine, GSK 3 beta peptide, v-akt murine thymoma viral oncogene homolog 1 (AKT1) | | Authors: | Morales, T.H, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of pyrrolopyrimidine inhibitors of Akt.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW3
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW4
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, GSK 3 beta peptide, RAC-alpha serine/threonine-protein kinase | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6U3I
 
 | |
6U2F
 
 | |
2P3W
 
 | |
1QTY
 
 | |
5HIB
 
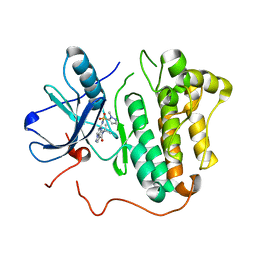 | | EGFR kinase domain mutant "TMLR" with a pyrazolopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-tert-butyl-5-{[(1-methyl-1H-pyrazol-5-yl)sulfonyl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HIC
 
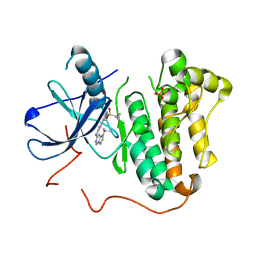 | | EGFR kinase domain mutant "TMLR" with a imidazopyridinyl-aminopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-{2-[1-(cyclopropylsulfonyl)-1H-pyrazol-4-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5KYC
 
 | |
5KYF
 
 | |
5KYB
 
 | |
5KYE
 
 | |
