-Search query
-Search result
Showing 1 - 50 of 1,792 items for (author: kim & y)

EMDB-18658: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex

PDB-8qu9: 
Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex
EMDB-36223: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp

PDB-8jg5: 
Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp
EMDB-43647: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant
EMDB-43650: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant

EMDB-43656: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)

EMDB-43657: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant (Locally refined map)

PDB-8vy7: 
CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant

PDB-8vy9: 
CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant

EMDB-43091: 
hGBP1 conformer on the bacterial outer membrane

EMDB-43153: 
hGBP1 conformer on the bacterial outer membrane

EMDB-37386: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP

EMDB-37387: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC1

EMDB-37388: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2

PDB-8w9z: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP

PDB-8wa0: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC1

PDB-8wa1: 
The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2
EMDB-39117: 
Cryo-EM structure of Maltose Binding Protein

PDB-8ybe: 
Cryo-EM structure of Maltose Binding Protein
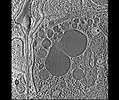
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
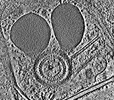
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
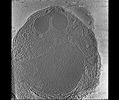
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite

EMDB-37465: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density

EMDB-37466: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE

PDB-8wdu: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density

PDB-8wdv: 
Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE
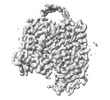
EMDB-35377: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)

EMDB-35378: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)

EMDB-35380: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex

EMDB-35382: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)

EMDB-35389: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)

EMDB-35390: 
Cryo-EM structure of G-protein free GPR156

PDB-8ieb: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)

PDB-8iec: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)

PDB-8ied: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex

PDB-8iei: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)

PDB-8iep: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)

PDB-8ieq: 
Cryo-EM structure of G-protein free GPR156

EMDB-35234: 
Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer

EMDB-35235: 
Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer

PDB-8i7v: 
Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer

PDB-8i7w: 
Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer

EMDB-35122: 
Human TRiC-PhLP2A complex in the closed state

EMDB-35199: 
The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state

PDB-8i1u: 
Human TRiC-PhLP2A complex in the closed state

PDB-8i6j: 
The focused refinement of CCT3-PhLP2A from TRiC-PhLP2A complex in the open state

EMDB-43049: 
Cryo-electron tomogram of mitochondria from cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43050: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43051: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
