[English] 日本語
 Yorodumi
Yorodumi- EMDB-36223: Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  NOROVIRUS / GI.4 / NOROVIRUS / GI.4 /  VIRUS LIKE PARTICLE / VIRUS LIKE PARTICLE /  human monoclonal antibody / Fv-clasp / human monoclonal antibody / Fv-clasp /  VIRUS VIRUS | |||||||||
| Biological species |   Norovirus / Norovirus /   Homo sapiens (human) Homo sapiens (human) | |||||||||
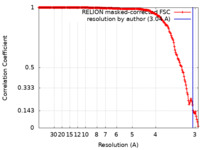
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.04 Å cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Hosaka T / Katsura K / Kimura-Someya T / Someya Y / Shirouzu M | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography reveal binding sites for human monoclonal antibodies Authors: Kimura-Someya T / Katsura K / Kato-Murayama M / Hosaka T / Uchikubo-Kamo T / Ihara K / Hanada K / Murayama K / Shirouzu M / Someya Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36223.map.gz emd_36223.map.gz | 449.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36223-v30.xml emd-36223-v30.xml emd-36223.xml emd-36223.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36223_fsc.xml emd_36223_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_36223.png emd_36223.png | 191.9 KB | ||
| Masks |  emd_36223_msk_1.map emd_36223_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36223.cif.gz emd-36223.cif.gz | 6 KB | ||
| Others |  emd_36223_additional_1.map.gz emd_36223_additional_1.map.gz emd_36223_half_map_1.map.gz emd_36223_half_map_1.map.gz emd_36223_half_map_2.map.gz emd_36223_half_map_2.map.gz | 13.2 MB 441.2 MB 441.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36223 http://ftp.pdbj.org/pub/emdb/structures/EMD-36223 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36223 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36223 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36223.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36223.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.47 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36223_msk_1.map emd_36223_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_36223_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36223_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36223_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Viral protein 1 with its antibody fragments
| Entire | Name: Viral protein 1 with its antibody fragments |
|---|---|
| Components |
|
-Supramolecule #1: Viral protein 1 with its antibody fragments
| Supramolecule | Name: Viral protein 1 with its antibody fragments / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Norovirus Norovirus |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Details: GenBank ID AB042808 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Norovirus Norovirus |
| Molecular weight | Theoretical: 58.108332 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASKDATPSA DGATGAGQLV PEVNTADPIP IDPVAGSSTA APVAGQVNLI DPWIINNFVQ APQGEFTISP NNTPGDVLFD LQLGPHLNP FLSHLSQMYN GWVGNMRVRV VLAGNAFTAG KVIICCVPPG FQSRTLSIAQ ATLFPHVIAD VRTLDPVEVP L EDVRNVLY ...String: MASKDATPSA DGATGAGQLV PEVNTADPIP IDPVAGSSTA APVAGQVNLI DPWIINNFVQ APQGEFTISP NNTPGDVLFD LQLGPHLNP FLSHLSQMYN GWVGNMRVRV VLAGNAFTAG KVIICCVPPG FQSRTLSIAQ ATLFPHVIAD VRTLDPVEVP L EDVRNVLY HNNDTQPTMR LLCMLYTPLR TGGASGGTDS FVVAGRVLTC PGPDFNFLFL VPPTVEQKTR PFTVPNIPLK YL SNSRIPN PIEGMSLSPD QTQNVQFQNG RCTIDGQPLG TTPVSVSQLC KFRGRITSGQ RVLNLTELDG SPFMAFAAPA PAG FPDLGS CDWHIEMSKI PNSSTQNNPI VTNSVKPNSQ QFVPHLSSIT LDENVSSGGD YIGTIQWTSP PSDSGGANTN FWKI PDYGS SLAEASQLAP AVYPPGFNEV IVYFMASIPG PNQSGSPNLV PCLLPQEYIT HFISEQAPIQ GEAALLHYVD PDTNR NLGE FKLYPGGYLT CVPNSSSTGP QQLPLDGVFV FASWVSRFYQ LKPVGTAGPA RGRLGVRR |
-Macromolecule #2: VH,SARAH
| Macromolecule | Name: VH,SARAH / type: protein_or_peptide / ID: 2 Details: VH-SARAH chimera of linker (residues (-6)-0), VH (residues 1-118), and SARAH (residues 119-171) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.847254 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSSGSSGEVQ LVESGAEVKK PGASVKVSCK ASGYTFTSLY MHWVRQAPGQ GLEWMGMINP SGGGTWNAQK FQGRVTMTRD TSTSTVYME LRSLRSDDTA MYYCARDSDQ YSQGLGYWGQ GTLVTVCSGS DYEFLKSWTV EDLQKRLLAL DPMMEQEIEE I RQKYQSKR QPILDAIEAK |
-Macromolecule #3: VL,SARAH
| Macromolecule | Name: VL,SARAH / type: protein_or_peptide / ID: 3 Details: VL-SARAH chimera of linker (residues (-6)-0), VL (residues 1-112), and SARAH (residues 113-162) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 18.19315 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: GSSGSSGQSA LTQPASVSGS PGQSITISCT GTSSDVGGYN YVSWYQQHPG KAPKLMIYDV SKRPSGVSNR FSGSKSGNTA SLTISGLQA KDEADYYCSS YTSSSTWVFG GGTKLTVLGG SDYEFLKSWT VEDLQKRLLA LDPMMEQEIE EIRQKYQCKR Q PILDAIEA K |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)