[English] 日本語
 Yorodumi
Yorodumi- EMDB-43650: CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 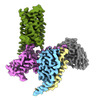 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  GPCR / GPCR /  Complex / Complex /  SIGNALING PROTEIN / SIGNALING PROTEIN /  Taste receptor Taste receptor | |||||||||
| Function / homology |  Function and homology information Function and homology informationbitter taste receptor activity /  taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor ...bitter taste receptor activity / taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor ...bitter taste receptor activity /  taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / taste receptor activity / detection of chemical stimulus involved in sensory perception of bitter taste / Class C/3 (Metabotropic glutamate/pheromone receptors) / G protein-coupled receptor activity / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane /  GTPase activity / GTPase activity /  synapse / protein-containing complex binding / synapse / protein-containing complex binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.88 Å cryo EM / Resolution: 2.88 Å | |||||||||
 Authors Authors | Kim Y / Gumpper RH / Roth BL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Bitter taste receptor activation by cholesterol and an intracellular tastant. Authors: Yoojoong Kim / Ryan H Gumpper / Yongfeng Liu / D Dewran Kocak / Yan Xiong / Can Cao / Zhijie Deng / Brian E Krumm / Manish K Jain / Shicheng Zhang / Jian Jin / Bryan L Roth /   Abstract: Bitter taste sensing is mediated by type 2 taste receptors (TAS2Rs (also known as T2Rs)), which represent a distinct class of G-protein-coupled receptors. Among the 26 members of the TAS2Rs, TAS2R14 ...Bitter taste sensing is mediated by type 2 taste receptors (TAS2Rs (also known as T2Rs)), which represent a distinct class of G-protein-coupled receptors. Among the 26 members of the TAS2Rs, TAS2R14 is highly expressed in extraoral tissues and mediates the responses to more than 100 structurally diverse tastants, although the molecular mechanisms for recognizing diverse chemicals and initiating cellular signalling are still poorly understood. Here we report two cryo-electron microscopy structures for TAS2R14 complexed with G (also known as gustducin) and G. Both structures have an orthosteric binding pocket occupied by endogenous cholesterol as well as an intracellular allosteric site bound by the bitter tastant cmpd28.1, including a direct interaction with the α5 helix of G and G. Computational and biochemical studies validate both ligand interactions. Our functional analysis identified cholesterol as an orthosteric agonist and the bitter tastant cmpd28.1 as a positive allosteric modulator with direct agonist activity at TAS2R14. Moreover, the orthosteric pocket is connected to the allosteric site via an elongated cavity, which has a hydrophobic core rich in aromatic residues. Our findings provide insights into the ligand recognition of bitter taste receptors and suggest activities of TAS2R14 beyond bitter taste perception via intracellular allosteric tastants. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43650.map.gz emd_43650.map.gz | 91 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43650-v30.xml emd-43650-v30.xml emd-43650.xml emd-43650.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43650.png emd_43650.png | 120.9 KB | ||
| Filedesc metadata |  emd-43650.cif.gz emd-43650.cif.gz | 7.2 KB | ||
| Others |  emd_43650_additional_1.map.gz emd_43650_additional_1.map.gz emd_43650_half_map_1.map.gz emd_43650_half_map_1.map.gz emd_43650_half_map_2.map.gz emd_43650_half_map_2.map.gz | 97 MB 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43650 http://ftp.pdbj.org/pub/emdb/structures/EMD-43650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43650 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43650 | HTTPS FTP |
-Related structure data
| Related structure data |  8vy9MC  8vy7C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43650.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43650.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.876 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_43650_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
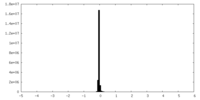
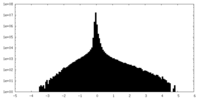
| Density Histograms |
-Half map: #2
| File | emd_43650_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
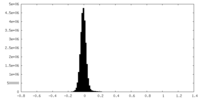
| Density Histograms |
-Half map: #1
| File | emd_43650_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
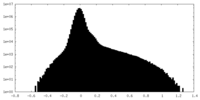
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of TAS2R14 with Ggust heterotrimer
| Entire | Name: Ternary complex of TAS2R14 with Ggust heterotrimer |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of TAS2R14 with Ggust heterotrimer
| Supramolecule | Name: Ternary complex of TAS2R14 with Ggust heterotrimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(t) subunit alpha-3
| Macromolecule | Name: Guanine nucleotide-binding protein G(t) subunit alpha-3 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.469027 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MGSTVSAEDK AAAERSKMID KNLREDAERD ARTVKLLLLG AGESGKATIV KQQMKIIHKN GYSEQECMEF KAVIYSNTLQ SILAIVKAM TTLGIDYVNP RSAEDQRQLY AMANTLEDGG MTPQLAEVIK RLWRDPGIQA CFERASEYQL NDSAAYYLND L DRITASGY ...String: MGSTVSAEDK AAAERSKMID KNLREDAERD ARTVKLLLLG AGESGKATIV KQQMKIIHKN GYSEQECMEF KAVIYSNTLQ SILAIVKAM TTLGIDYVNP RSAEDQRQLY AMANTLEDGG MTPQLAEVIK RLWRDPGIQA CFERASEYQL NDSAAYYLND L DRITASGY VPNEQDVLHS RVKTTGIIET QFSFKDLHFR MFDVGAQRSE RKKWIHCFEG VTCIIFCAAL SAYDMVLVED EE VNRMHAS LKLFDSICNH KYFSDTSIVL FLNKKDIFQE KVTKVHLSIC FPEYTGPNTF EDAGNYIKNQ FLDLNLKKED KEI YSHMTC STDTQNVKFV FDAVTDIIIK ENLKDCGLF |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.936668 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WNGGSGGGGS GGSSSGGGGS GGGGSGGSSS GGVSGWRLFK KISG GS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 28.668922 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAALEVLFQ GPHHHHHHHH |
-Macromolecule #5: Taste receptor type 2 member 14,GPCR,Taste receptor type 2 member 14
| Macromolecule | Name: Taste receptor type 2 member 14,GPCR,Taste receptor type 2 member 14 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 72.722641 KDa |
| Recombinant expression | Organism:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
| Sequence | String: DYKDDDDAKL QTMHHHHHHH HHHENLYFQG GTTMADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LGSTLEVLFQ GPMGGVIKSI F TFVLIVEF ...String: DYKDDDDAKL QTMHHHHHHH HHHENLYFQG GTTMADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LGSTLEVLFQ GPMGGVIKSI F TFVLIVEF IIGNLGNSFI ALVNCIDWVK GRKISSVDRI LTALAISRIS LVWLIFGSWC VSVFFPALFA TEKMFRMLTN IW TVINHFS VWLATGLGTF YFLKIANFSN SIFLYLKWRV KKVVLVLLLV TSVFLFLNIA LINIHINASI NGYRRNKTCS SDS SNFTRF SSLIVLTSTV FIFIPFTLSL AMFLLLIFSM WKHRKKMQHT VKISGDASTR AHRGVKSVIT FFLLYAIFSL SFFI SVWTS ERLEENLIIL SQVMGMAYPS CHSCVLILGN KKLRQASLSV LLWLRYMFKD GEPSGHKEFR ESSGSAGSAG SGGSG GGSG GGGSGGSSSG GVFTLEDFVG DWEQTAAYNL DQVLEQGGVS SLLQNLAVSV TPIQRIVRSG ENALKIDIHV IIPYEG LSA DQMAQIEEVF KVVYPVDDHH FKVILPYGTL VIDGVTPNML NYFGRPYEGI AVFDGKKITV TGTLWNGNKI IDERLIT PD GSMLFRVTIN S UniProtKB: Taste receptor type 2 member 14 |
-Macromolecule #6: 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)p...
| Macromolecule | Name: 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine type: ligand / ID: 6 / Number of copies: 1 / Formula: A1AEI |
|---|---|
| Molecular weight | Theoretical: 321.261 Da |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source: OTHER |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 620305 |
|---|---|
| Startup model | Type of model: NONE |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: OTHER |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.88 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 4.2) / Number images used: 458284 |
 Movie
Movie Controller
Controller























 Z
Z Y
Y X
X