+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GPR156-miniGo-scFv16 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Membrane protein / Membrane protein /  G-protein coupled receptor / G-protein coupled receptor /  Signal transduction / Signal transduction /  Phospholipid Phospholipid | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / mu-type opioid receptor binding /  corticotropin-releasing hormone receptor 1 binding / gamma-aminobutyric acid signaling pathway / dopamine receptor signaling pathway / G protein-coupled serotonin receptor binding / corticotropin-releasing hormone receptor 1 binding / gamma-aminobutyric acid signaling pathway / dopamine receptor signaling pathway / G protein-coupled serotonin receptor binding /  muscle contraction / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway ...G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / mu-type opioid receptor binding / muscle contraction / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway ...G protein-coupled GABA receptor activity / G protein-coupled receptor heterodimeric complex / mu-type opioid receptor binding /  corticotropin-releasing hormone receptor 1 binding / gamma-aminobutyric acid signaling pathway / dopamine receptor signaling pathway / G protein-coupled serotonin receptor binding / corticotropin-releasing hormone receptor 1 binding / gamma-aminobutyric acid signaling pathway / dopamine receptor signaling pathway / G protein-coupled serotonin receptor binding /  muscle contraction / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / muscle contraction / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade /  extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / G alpha (12/13) signalling events / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane /  GTPase activity / GTPase activity /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  membrane / membrane /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.33 Å cryo EM / Resolution: 3.33 Å | |||||||||
 Authors Authors | Shin J / Park J / Cho Y | |||||||||
| Funding support |  Korea, Republic Of, 1 items Korea, Republic Of, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Constitutive activation mechanism of a class C GPCR. Authors: Jinwoo Shin / Junhyeon Park / Jieun Jeong / Jordy Homing Lam / Xingyu Qiu / Di Wu / Kuglae Kim / Joo-Youn Lee / Carol V Robinson / Jaekyung Hyun / Vsevolod Katritch / Kwang Pyo Kim / Yunje Cho /    Abstract: Class C G-protein-coupled receptors (GPCRs) are activated through binding of agonists to the large extracellular domain (ECD) followed by rearrangement of the transmembrane domains (TMDs). GPR156, a ...Class C G-protein-coupled receptors (GPCRs) are activated through binding of agonists to the large extracellular domain (ECD) followed by rearrangement of the transmembrane domains (TMDs). GPR156, a class C orphan GPCR, is unique because it lacks an ECD and exhibits constitutive activity. Impaired GPR156-G signaling contributes to loss of hearing. Here we present the cryo-electron microscopy structures of human GPR156 in the G-free and G-coupled states. We found that an endogenous phospholipid molecule is located within each TMD of the GPR156 dimer. Asymmetric binding of Gα to the phospholipid-bound GPR156 dimer restructures the first and second intracellular loops and the carboxy-terminal part of the elongated transmembrane 7 (TM7) without altering dimer conformation. Our findings reveal that GPR156 is a transducer for phospholipid signaling. Constant binding of abundant phospholipid molecules and the G-protein-induced reshaping of the cytoplasmic face provide a basis for the constitutive activation of GPR156. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35380.map.gz emd_35380.map.gz | 248.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35380-v30.xml emd-35380-v30.xml emd-35380.xml emd-35380.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35380.png emd_35380.png | 127.6 KB | ||
| Filedesc metadata |  emd-35380.cif.gz emd-35380.cif.gz | 6.7 KB | ||
| Others |  emd_35380_half_map_1.map.gz emd_35380_half_map_1.map.gz emd_35380_half_map_2.map.gz emd_35380_half_map_2.map.gz | 243.8 MB 243.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35380 http://ftp.pdbj.org/pub/emdb/structures/EMD-35380 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35380 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35380 | HTTPS FTP |
-Related structure data
| Related structure data |  8iedMC  8iebC  8iecC  8ieiC  8iepC  8ieqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35380.map.gz / Format: CCP4 / Size: 262.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35380.map.gz / Format: CCP4 / Size: 262.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35380_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
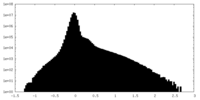
| Projections & Slices |
| ||||||||||||
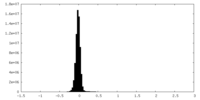
| Density Histograms |
-Half map: #1
| File | emd_35380_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
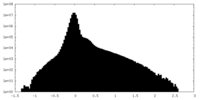
| Projections & Slices |
| ||||||||||||
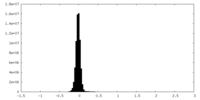
| Density Histograms |
- Sample components
Sample components
+Entire : GPR156-miniGo-scFv16 complex
+Supramolecule #1: GPR156-miniGo-scFv16 complex
+Supramolecule #2: GPR156
+Supramolecule #3: miniGo heterotrimeric complex
+Supramolecule #4: scFv16
+Macromolecule #1: Probable G-protein coupled receptor 156
+Macromolecule #2: Guanine nucleotide-binding protein G(o) subunit alpha
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #4: Single-chain variable fragment scFv16
+Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #6: [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(tri...
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER Details: GPR156: AlphaFold2 G-protein and scFv16: 7UM5 (PDB) |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.33 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 205728 |
 Movie
Movie Controller
Controller




























 Z
Z Y
Y X
X