2K0R
 
 | | Solution structure of the C103S mutant of the N-terminal Domain of DsbD from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Quinternet, M, Selme, L, Tsan, P, Beaufils, C, Jacob, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-11-11 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the cysteine 103 to serine mutant of the N-terminal domain of DsbD from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2K9F
 
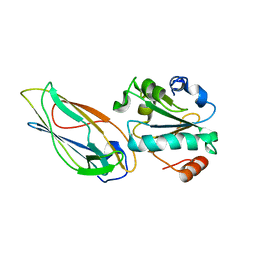 | | Structural features of the complex between the DsbD N-terminal and the PilB N-terminal domains from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein dsbD, Thioredoxin | | Authors: | Quinternet, M, Tsan, P, Selme, L, Jacob, C, Boschi-Muller, S, Branlant, G, Cung, M. | | Deposit date: | 2008-10-09 | | Release date: | 2009-05-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Formation of the complex between DsbD and PilB N-terminal domains from Neisseria meningitidis necessitates an adaptability of nDsbD.
Structure, 17, 2009
|
|
6TRV
 
 | |
7JST
 
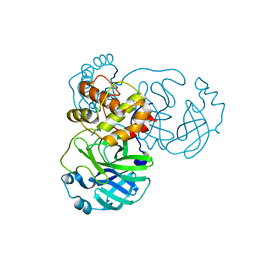 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
5KQA
 
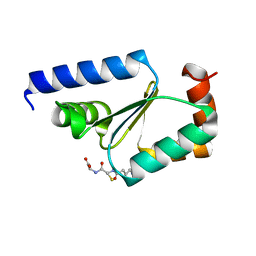 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
5LLK
 
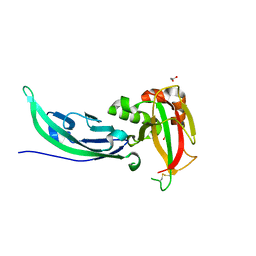 | | Crystal structure of human alpha-dystroglycan | | Descriptor: | 1,2-ETHANEDIOL, Dystroglycan | | Authors: | Covaceuszach, S, Cassetta, A, Lamba, D, Brancaccio, A, Bozzi, M, Sciandra, F, Bigotti, M.G, Konarev, P.V. | | Deposit date: | 2016-07-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility of human alpha-dystroglycan.
FEBS Open Bio, 7, 2017
|
|
8KGZ
 
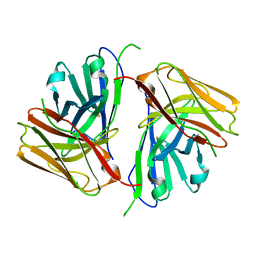 | | Crystal structure of single-chain Fv antibody against antigen peptide from SARS-CoV2 S-spike protein | | Descriptor: | Thioredoxin 1,scFv | | Authors: | Ma, Q.Q, Zhang, B.L, Cheng, X.Y, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2023-08-20 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of sheep scFv antibody against an antigen peptide of SARS-CoV2 S-spike protein
To Be Published
|
|
8T4C
 
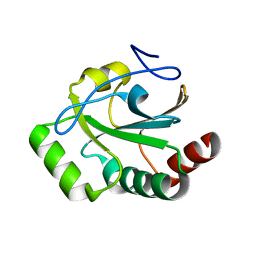 | |
6XAR
 
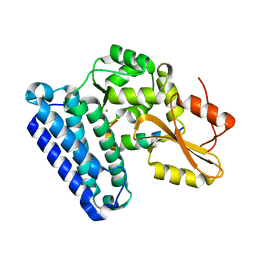 | |
6Y4Z
 
 | | The crystal structure of human MACROD2 in space group P43212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6Y4Y
 
 | | The crystal structure of human MACROD2 in space group P41212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5CJP
 
 | | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs | | Descriptor: | Cell division cycle 42 (GTP binding protein, 25kDa), isoform CRA_a, ... | | Authors: | Worthylake, D.K, Boyapati, V.K, LeCour Jr, L. | | Deposit date: | 2015-07-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis for Cdc42-Induced Dimerization of IQGAPs.
Structure, 24, 2016
|
|
5E4W
 
 | | Crystal structure of cpSRP43 chromodomains 2 and 3 in complex with the Alb3 tail | | Descriptor: | CALCIUM ION, GLYCEROL, Inner membrane protein ALBINO3, ... | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
5EZU
 
 | | Crystal structure of the N-terminal domain of vaccinia virus immunomodulator A46 in complex with myristic acid. | | Descriptor: | MYRISTIC ACID, Protein A46 | | Authors: | Fedosyuk, S, Bezerra, G.A, Sammito, M, Uson, I, Skern, T. | | Deposit date: | 2015-11-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Vaccinia Virus Immunomodulator A46: A Lipid and Protein-Binding Scaffold for Sequestering Host TIR-Domain Proteins.
PLoS Pathog., 12, 2016
|
|
4V2L
 
 | |
7TKV
 
 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-17 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
4V2N
 
 | |
4V2M
 
 | |
4X43
 
 | |
7OOL
 
 | | Crystal structure of a Candidatus photodesmus katoptron thioredoxin chimera containing an ancestral loop | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gavira, J.A, Ibarra-Molero, B, Gamiz-Arco, G, Risso, V, Sanchez-Ruiz, J.M. | | Deposit date: | 2021-05-28 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining Ancestral Reconstruction with Folding-Landscape Simulations to Engineer Heterologous Protein Expression.
J.Mol.Biol., 433, 2021
|
|
7V8H
 
 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XPW
 
 | | Structure of OhTRP14 | | Descriptor: | Thioredoxin domain-containing protein 17 | | Authors: | Wang, S.Q, Huang, S.Q. | | Deposit date: | 2022-05-05 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into the redox regulation of Oncomelania hupensis TRP14 and its potential role in the snail host response to parasite invasion.
Fish Shellfish Immunol., 128, 2022
|
|
7XQ3
 
 | |
3AQX
 
 | | Crystal structure of Bombyx mori beta-GRP/GNBP3 N-terminal domain with laminarihexaoses | | Descriptor: | Beta-1,3-glucan-binding protein, GLYCEROL, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
3AQY
 
 | | Crystal structure of Plodia interpunctella beta-GRP/GNBP3 N-terminal domain | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Kanagawa, M, Satoh, T, Ikeda, A, Adachi, Y, Ohno, N, Yamaguchi, Y. | | Deposit date: | 2010-11-22 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor
J.Biol.Chem., 286, 2011
|
|
