5AY3
 
 | | Crystal structure of RNA duplex containing C-C base pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*A*CP*UP*CP*C)-3') | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4JRT
 
 | | Crystal structure of an A-form RNA duplex containing three GU base pairs | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*CP*AP*CP*UP*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*UP*GP*GP*UP*GP*CP*GP*GP*G)-3') | | Authors: | Kondo, J, Dock-Bregeon, A.C, Willkomm, D.K, Hartmann, R.K, Westhof, E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an A-form RNA duplex obtained by degradation of 6S RNA in a crystallization droplet
Acta Crystallogr.,Sect.F, 69, 2013
|
|
8FEP
 
 | |
8FER
 
 | |
6Z18
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | Descriptor: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | Authors: | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
5UX3
 
 | |
7DD4
 
 | |
6XUR
 
 | | RNA dodecamer with a 6-hydrazino-2-aminopurine modified base | | Descriptor: | MAGNESIUM ION, RNA dodecamer with a 6-hydrazino-2-aminopurine modified base | | Authors: | Ennifar, E, Micura, R, Gasser, C, Brillet, K. | | Deposit date: | 2020-01-21 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thioguanosine Conversion Enables mRNA-Lifetime Evaluation by RNA Sequencing Using Double Metabolic Labeling (TUC-seq DUAL).
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XUS
 
 | | RNA dodecamer with a 6-hydrazino-2-aminopurine modified base | | Descriptor: | MAGNESIUM ION, RNA dodecamer with a 6-hydrazino-2-aminopurine modified base, SODIUM ION | | Authors: | Ennifar, E, Micura, R, Gasser, C, Brillet, K. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thioguanosine Conversion Enables mRNA-Lifetime Evaluation by RNA Sequencing Using Double Metabolic Labeling (TUC-seq DUAL).
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1M82
 
 | | SOLUTION STRUCTURE OF THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Descriptor: | RNA (25-MER): THE COMPLEMENTARY RNA PROMOTER OF INFLUENZA A VIRUS | | Authors: | Park, C.-J, Bae, S.-H, Lee, M.-K, Varani, G, Choi, B.-S. | | Deposit date: | 2002-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
NUCLEIC ACIDS RES., 31, 2003
|
|
5HN2
 
 | |
5HNJ
 
 | |
7JRS
 
 | |
7JRT
 
 | |
5X3Z
 
 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
1KOC
 
 | | RNA APTAMER COMPLEXED WITH ARGININE, NMR | | Descriptor: | ARGININE, RNA (5'-R(P*AP*CP*AP*GP*GP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*CP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
5V0H
 
 | | RNA duplex with 2-MeImpG analogue bound-one binding site | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*G)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
1KOD
 
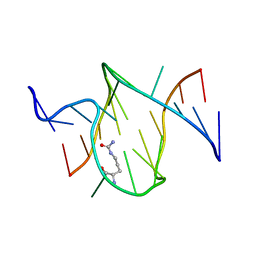 | | RNA APTAMER COMPLEXED WITH CITRULLINE, NMR | | Descriptor: | CITRULLINE, RNA (5'-R(P*AP*CP*GP*GP*UP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*UP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
4QI2
 
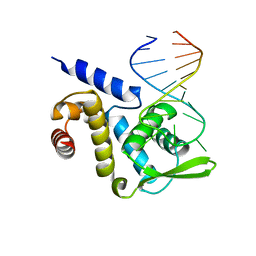 | | X-ray structure of the ROQ domain from murine Roquin-1 in complex with a 23-mer Tnf-CDE RNA | | Descriptor: | RNA (5'-R(*AP*CP*AP*UP*GP*UP*UP*UP*UP*CP*UP*GP*UP*GP*AP*AP*AP*AP*CP*GP*GP*AP*G)-3'), Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5V0J
 
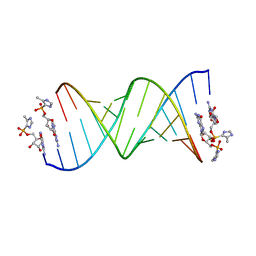 | | RNA duplex with 2-MeImpG analogue bound-2 binding sites | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
1NBR
 
 | |
466D
 
 | | DISORDER AND TWIN REFINEMENT OF RNA HEPTAMER DOUBLE HELIX | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | Authors: | Mueller, U, Muller, Y.A, Herbst-Irmer, R, Sprinzl, M, Heinemann, U. | | Deposit date: | 1999-04-14 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Disorder and twin refinement of RNA heptamer double helices.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1FHK
 
 | |
5C5W
 
 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
472D
 
 | | STRUCTURE OF AN OCTAMER RNA WITH TANDEM GG/UU MISPAIRS | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*GP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*UP*UP*UP*AP*C)-3') | | Authors: | Deng, J, Sundaralingam, M. | | Deposit date: | 1999-05-14 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G.G/U.U tandem wobble base pairs: comparison with other tandem G.U pairs.
Nucleic Acids Res., 28, 2000
|
|
