4F0U
 
 | |
4F0T
 
 | |
4GXE
 
 | | T. vulcanus Phycocyanin crystallized in 4M Urea | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Marx, A, Adir, N. | | Deposit date: | 2012-09-04 | | Release date: | 2013-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly.
Biochim.Biophys.Acta, 1827, 2013
|
|
4H0M
 
 | |
4GY3
 
 | | T. vulcanus Phycocyanin crystallized in 2M Urea | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Marx, A, Adir, N. | | Deposit date: | 2012-09-05 | | Release date: | 2013-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allophycocyanin and phycocyanin crystal structures reveal facets of phycobilisome assembly.
Biochim.Biophys.Acta, 1827, 2013
|
|
2B3I
 
 | | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (19 STRUCTURES) | | Descriptor: | COPPER (I) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Babu, C.R, Volkman, B.F, Bullerjahn, G.S. | | Deposit date: | 1998-12-11 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica.
Biochemistry, 38, 1999
|
|
3BG5
 
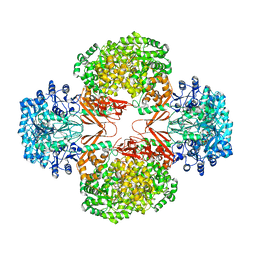 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5J4D
 
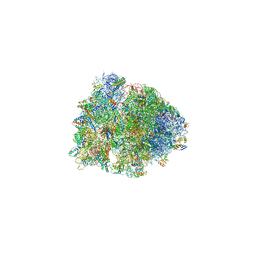 | |
6HRN
 
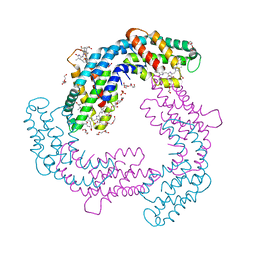 | | C-Phycocyanin from heterocyst forming filamentous cyanobacterium Nostoc sp. WR13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha Subunit of Cyanobacterial Phycocyanin protein, Beta Subunit of Cyanobacterial Phycocyanin protein, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2018-09-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.513 Å) | | Cite: | Crystal structure of phycocyanin from heterocyst-forming filamentous cyanobacterium Nostoc sp. WR13.
Int.J.Biol.Macromol., 135, 2019
|
|
1F99
 
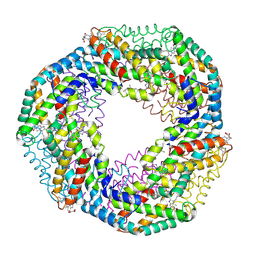 | | CRYSTAL STRUCTURE OF R-PHYCOCYANIN FROM POLYSIPHONIA AT 2.4 A RESOLUTION | | Descriptor: | BILIVERDINE IX ALPHA, PHYCOCYANOBILIN, PHYCOERYTHROBILIN, ... | | Authors: | Liang, D.C, Jiang, T, Chang, W.R. | | Deposit date: | 2000-07-09 | | Release date: | 2001-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of R-phycocyanin and possible energy transfer pathways in the phycobilisome.
Biophys.J., 81, 2001
|
|
3NPH
 
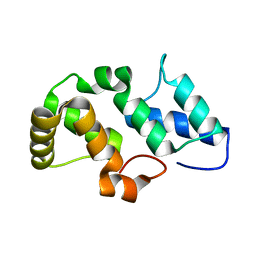 | | Crystal structure of the pfam00427 domain from Synechocystis sp. PCC 6803 | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod 2 | | Authors: | Gao, X, Chen, L, Wu, J.-W, Zhang, Y.-Z. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Crystal structure of the N-terminal domain of linker L(R) and the assembly of cyanobacterial phycobilisome rods
Mol.Microbiol., 82, 2011
|
|
1KN6
 
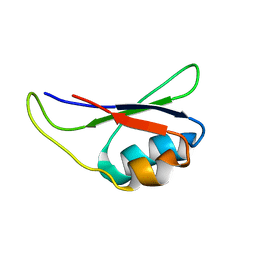 | |
4M6V
 
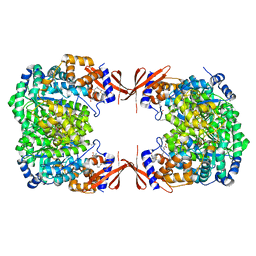 | |
4LOC
 
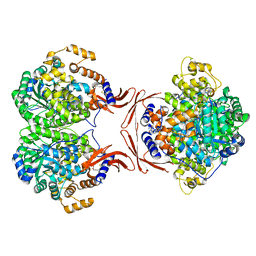 | |
2AF9
 
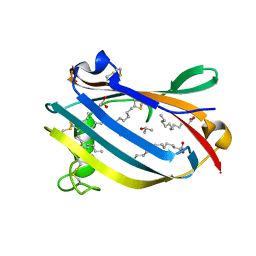 | | Crystal Structure analysis of GM2-Activator protein complexed with phosphatidylcholine | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, LAURIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG2
 
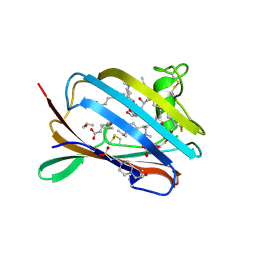 | | Crystal Structure Analysis of GM2-activator protein complexed with Phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG9
 
 | | Crystal Structure of the Y137S mutant of GM2-Activator Protein | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG4
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, Ganglioside GM2 activator, ISOPROPYL ALCOHOL, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2HXA
 
 | |
2HX9
 
 | |
2HX7
 
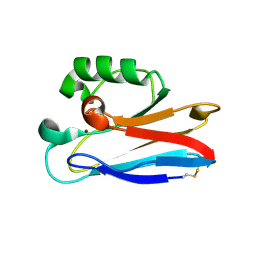 | |
2HX8
 
 | |
3TW6
 
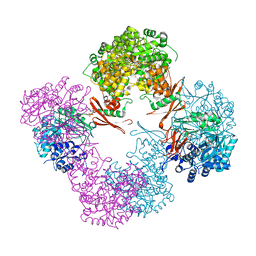 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
3TW7
 
 | | Structure of Rhizobium etli pyruvate carboxylase T882A crystallized without acetyl coenzyme-A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pyruvate carboxylase protein, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
1A9X
 
 | |
