5Z9F
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9L
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 2-fluoro-4-hydroxybenzonitrile, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9Q
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 5-phenyl-1H-pyrazol-3-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z4O
 
 | |
5Z9B
 
 | | Bacterial GyrB ATPase domain in complex with (3,4-dichlorophenyl)hydrazine | | Descriptor: | (3,4-dichlorophenyl)hydrazine, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-02-02 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
5Z9M
 
 | |
5Z4H
 
 | | Bacterial GyrB ATPase domain in complex with a chemical fragment | | Descriptor: | 1H-benzimidazol-2-amine, 4-(4-hydroxyphenyl)sulfanylphenol, DNA gyrase subunit B, ... | | Authors: | Huang, X, Zhou, H. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an auxiliary druggable pocket in the DNA gyrase ATPase domain using fragment probes
Medchemcomm, 9, 2018
|
|
6F96
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6CJI
 
 | | Candida albicans Hsp90 nucleotide binding domain | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJR
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with SNX-2112 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein 90 homolog | | Authors: | Kirkpatrick, M.G, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJJ
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90 homolog, ... | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, STRUCTURAL GENOMICS CONSORTIUM, S.G.C, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJS
 
 | |
6CJP
 
 | | Candida albicans Hsp90 nucleotide binding domain in complex with radicicol | | Descriptor: | (1aR,2Z,4E,6E,14R,15aR)-9,11-dihydroxy-6-{[(4-methoxyphenyl)methoxy]imino}-14-methyl-1a,6,7,14,15,15a-hexahydro-12H-oxireno[e][2]benzoxacyclotetradecin-12-one, Heat shock protein 90 homolog | | Authors: | Nation, C, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
6CJL
 
 | |
6GAV
 
 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B,DNA gyrase subunit A | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, MAyer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
6GAU
 
 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | DNA gyrase subunit B,DNA gyrase subunit A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, Mayer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
6DK7
 
 | | RetS histidine kinase region with cobalt | | Descriptor: | COBALT (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6DK8
 
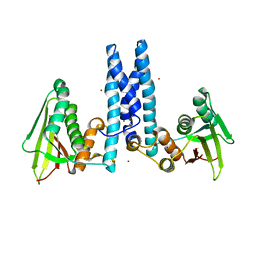 | | RetS kinase region without cobalt | | Descriptor: | NICKEL (II) ION, RetS (Regulator of Exopolysaccharide and Type III Secretion) | | Authors: | Mancl, J.M, Schubot, F.D. | | Deposit date: | 2018-05-29 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Helix Cracking Regulates the Critical Interaction between RetS and GacS in Pseudomonas aeruginosa.
Structure, 27, 2019
|
|
6ENG
 
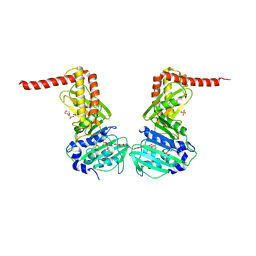 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
6ENH
 
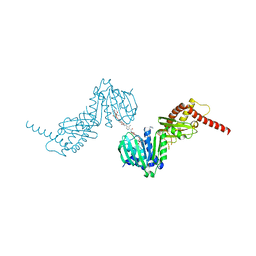 | |
5ZXM
 
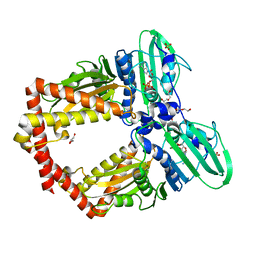 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F86
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6RKW
 
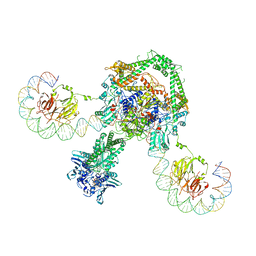 | | CryoEM structure of the complete E. coli DNA Gyrase complex bound to a 130 bp DNA duplex | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (58-MER), DNA (62-MER), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
