3RFB
 
 | | Structure of fRMsr | | Descriptor: | METHIONINE SULFOXIDE, Putative uncharacterized protein | | Authors: | Bong, S.M, Chi, Y.M. | | Deposit date: | 2011-04-06 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of fRMsr
to be published
|
|
3S7Q
 
 | | Crystal Structure of a Monomeric Infrared Fluorescent Deinococcus radiodurans Bacteriophytochrome chromophore binding domain | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Auldridge, M.E, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2011-05-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
3S7O
 
 | | Crystal Structure of the Infrared Fluorescent D207H variant of Deinococcus Bacteriophytochrome chromophore binding domain at 1.24 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, GLYCEROL | | Authors: | Forest, K.T, Auldridge, M.E, Satyshur, K.A, Anstrom, D.M. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
3S7N
 
 | |
3S7P
 
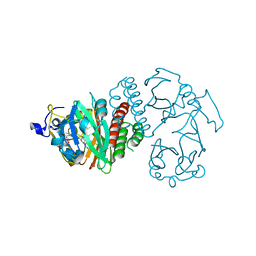 | |
3NHQ
 
 | | The dark Pfr structure of the photosensory core module of P. aeruginosa Bacteriophytochrome | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Yang, X, Ren, Z, Kuk, J, Moffat, K. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome.
Nature, 479, 2011
|
|
3ZQ5
 
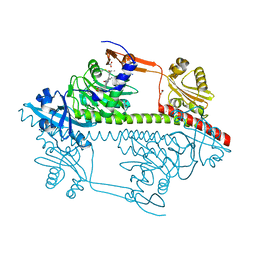 | | Structure of the Y263F mutant of the cyanobacterial phytochrome Cph1 | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mailliet, J, Psakis, G, Sineshchekov, V, Essen, L.-O, Hughes, J. | | Deposit date: | 2011-06-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Spectroscopy and a High-Resolution Crystal Structure of Tyr263 Mutants of Cyanobacterial Phytochrome Cph1.
J.Mol.Biol., 413, 2011
|
|
3TRC
 
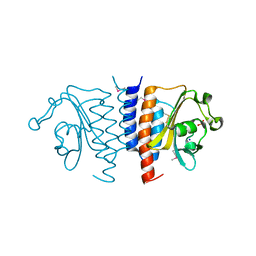 | | Structure of the GAF domain from a phosphoenolpyruvate-protein phosphotransferase (ptsP) from Coxiella burnetii | | Descriptor: | PHOSPHATE ION, Phosphoenolpyruvate-protein phosphotransferase, SODIUM ION | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
2LB9
 
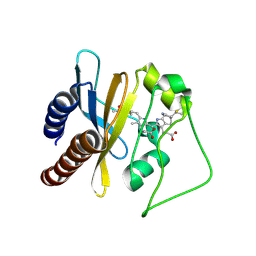 | | Refined solution structure of a cyanobacterial phytochrome gaf domain in the red light-absorbing ground state (corrected pyrrole ring planarity) | | Descriptor: | PHYCOCYANOBILIN, Sensor histidine kinase | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state.
J.Mol.Biol., 383, 2008
|
|
2LB5
 
 | | Refined Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor histidine kinase | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form.
Nature, 463, 2010
|
|
2XSS
 
 | |
2Y8H
 
 | |
2Y79
 
 | | STRUCTURE OF THE FIRST GAF DOMAIN E87A MUTANT OF MYCOBACTERIUM TUBERCULOSIS DOSS | | Descriptor: | CALCIUM ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cho, H.Y, Cho, H.J, Kang, B.S. | | Deposit date: | 2011-01-30 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Blockage of the Channel to Heme by the E87 Side Chain in the Gaf Domain of Mycobacterium Tuberculosis Doss Confers the Unique Sensitivity of Doss to Oxygen.
FEBS Lett., 585, 2011
|
|
3KO6
 
 | |
3P01
 
 | |
3LFV
 
 | | crystal structure of unliganded PDE5A GAF domain | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
3MF0
 
 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
3O5Y
 
 | |
3K2N
 
 | |
2KLI
 
 | | Structural Basis for the Photoconversion of A Phytochrome to the Activated FAR-RED LIGHT-ABSORBING Form | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Zhang, J, Rivera, M, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-03 | | Release date: | 2009-11-03 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
2KOI
 
 | | Refined solution structure of a cyanobacterial phytochrome GAF domain in the red light-absorbing ground state | | Descriptor: | PHYCOCYANOBILIN, Sensor protein | | Authors: | Cornilescu, C.C, Cornilescu, G, Ulijasz, A.T, Vierstra, R.D, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-09-22 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the photoconversion of a phytochrome to the activated Pfr form
Nature, 463, 2010
|
|
3IBJ
 
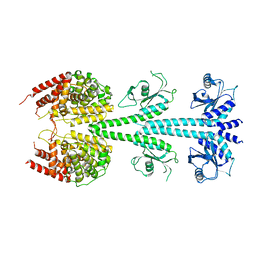 | | X-ray structure of PDE2A | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-dependent 3',5'-cyclic phosphodiesterase | | Authors: | Pandit, J. | | Deposit date: | 2009-07-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G6O
 
 | |
3IBR
 
 | |
