8BC3
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, BmSF-TAL | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8C4I
 
 | |
3HJZ
 
 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1F05
 
 | |
6YR3
 
 | |
6YRH
 
 | |
6YS0
 
 | |
6YRM
 
 | |
6YRT
 
 | |
6YRE
 
 | | Transaldolase variant T30C/D211C from T. acidophilum | | Descriptor: | ACETATE ION, GLYCEROL, Probable transaldolase | | Authors: | Sautner, V, Lietzow, T.H, Tittmann, K. | | Deposit date: | 2020-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Large-scale motions underlie physical but not chemical steps in transaldolase mechanism: Substrate binding by conformational selection and rate-determining product release
To Be Published
|
|
1I2Q
 
 | |
1I2O
 
 | |
1I2N
 
 | |
1I2P
 
 | |
1I2R
 
 | |
6ZWJ
 
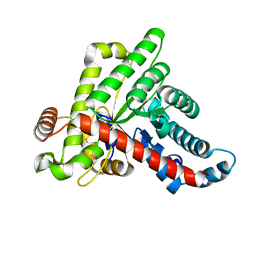 | | Neisseria gonorrhoeae transaldolase at 1.35 Angstrom resolution | | Descriptor: | CITRIC ACID, GLYCEROL, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZWF
 
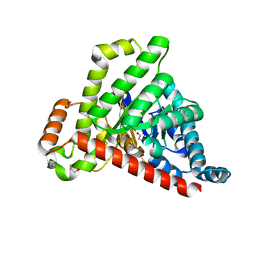 | | Neisseria gonorrhoeae transaldolase | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CITRIC ACID, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.-M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZX4
 
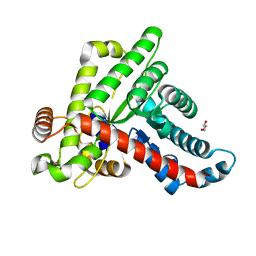 | | Neisseria gonorrhoeae transaldolase | | Descriptor: | CITRIC ACID, GLYCEROL, Transaldolase | | Authors: | Sautner, V, Rabe von Pappenheim, F, Wensien, M, Tittmann, K. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
6ZWH
 
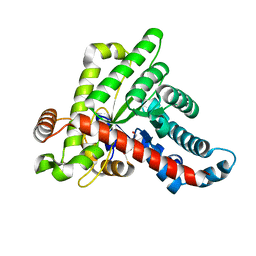 | | Neisseria gonorrhoeae transaldolase at 1.5 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.-M, Sautner, V, Tittmann, K. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
7B0L
 
 | |
7BBW
 
 | |
7BBX
 
 | | Neisseria gonorrhoeae transaldolase, variant K8A | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Transaldolase | | Authors: | Rabe von Pappenheim, F, Wensien, M, Funk, L.M, Tittmann, K. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
4RXG
 
 | | Fructose-6-phosphate aldolase Q59E from E.coli | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL, PENTAETHYLENE GLYCOL | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
4S1F
 
 | | Fructose-6-phosphate aldolase A from E.coli soaked in acetylacetone | | Descriptor: | Fructose-6-phosphate aldolase 1, pentane-2,4-dione | | Authors: | Stellmacher, L, Sandalova, T, Leptihn, S, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-13 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Acid Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase
ChemCatChem, 2015
|
|
4S2C
 
 | | Covalent complex of E. coli transaldolase TalB with fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, FRUCTOSE -6-PHOSPHATE, Transaldolase B | | Authors: | Stellmacher, L, Sandalova, T, Schneider, G, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel mode of inhibition by D-tagatose 6-phosphate through a Heyns rearrangement in the active site of transaldolase B variants.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
