1WCQ
 
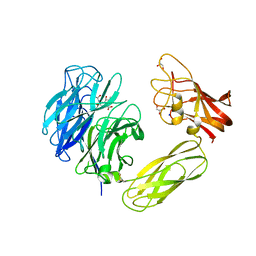 | | Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE, ... | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-11-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Nucleophilic Mutants of the Micromonospora Viridifaciens Sialidase Operate with Retention of Configuration by Two Different Mechanisms.
Chembiochem, 6, 2005
|
|
5ZU5
 
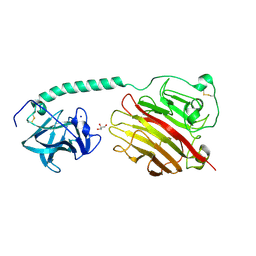 | |
4ZRX
 
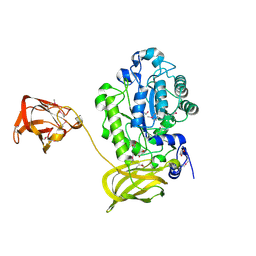 | |
2BER
 
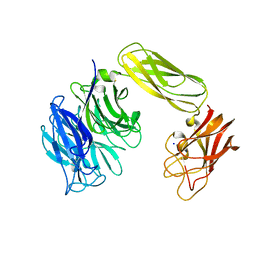 | | Y370G Active Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). | | Descriptor: | BACTERIAL SIALIDASE, N-acetyl-beta-neuraminic acid, SODIUM ION | | Authors: | Newstead, S, Watson, J.N, Bennet, A.J, Taylor, G.L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of Action of an Inverting Mutant Sialidase.
Biochemistry, 44, 2005
|
|
2BZD
 
 | |
5K8D
 
 | | Crystal structure of rFVIIIFc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Leksa, N, Quan, C. | | Deposit date: | 2016-05-29 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | The structural basis for the functional comparability of factor VIII and the long-acting variant recombinant factor VIII Fc fusion protein.
J. Thromb. Haemost., 15, 2017
|
|
5K9H
 
 | | Crystal structure of a glycoside hydrolase 29 family member from an unknown rumen bacterium | | Descriptor: | 0940_GH29, GLYCEROL, SODIUM ION, ... | | Authors: | Summers, E.L, Arcus, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The structure of a glycoside hydrolase 29 family member from a rumen bacterium reveals unique, dual carbohydrate-binding domains.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1SDD
 
 | | Crystal Structure of Bovine Factor Vai | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Adams, T.E, Hockin, M.F, Mann, K.G, Everse, S.J. | | Deposit date: | 2004-02-13 | | Release date: | 2004-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of activated protein C-inactivated bovine factor Va: Implications for cofactor function.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2ZXQ
 
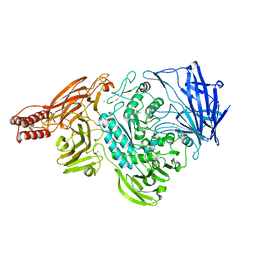 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
7W12
 
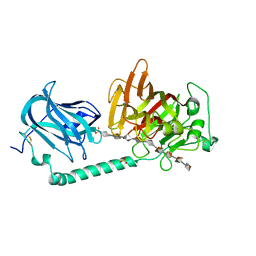 | | Complex structure of alginate lyase AlyB-OU02 with G9 | | Descriptor: | Alginate lyase, SULFATE ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
1T2X
 
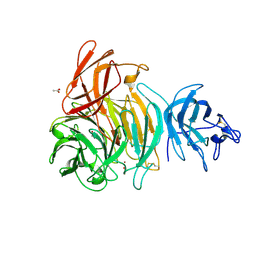 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
5MSX
 
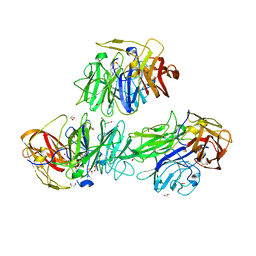 | | Glycoside hydrolase BT_3662 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
3CDZ
 
 | | Crystal structure of human factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ngo, J.C, Huang, M, Roth, D.A, Furie, B.C, Furie, B. | | Deposit date: | 2008-02-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex.
Structure, 16, 2008
|
|
1EUU
 
 | | SIALIDASE OR NEURAMINIDASE, LARGE 68KD FORM | | Descriptor: | SIALIDASE, SODIUM ION, beta-D-galactopyranose | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
1W8O
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens | | Descriptor: | BACTERIAL SIALIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
1W8N
 
 | | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, BACTERIAL SIALIDASE, SODIUM ION, ... | | Authors: | Newstead, S, Watson, J.N, Dookhun, V, Bennet, A.J, Taylor, G. | | Deposit date: | 2004-09-24 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora Viridifaciens
FEBS Lett., 577, 2004
|
|
6SH9
 
 | | EngBF DARPin Fusion 4b D12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-alpha-N-acetylgalactosaminidase,DARPin 4b D12, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-08-06 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6TR3
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 in complex with fucose | | Descriptor: | CALCIUM ION, F5/8 type C domain-containing protein, MAGNESIUM ION, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
6TR4
 
 | | Ruminococcus gnavus GH29 fucosidase E1_10125 D221A mutant in complex with fucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, F5/8 type C domain-containing protein, ... | | Authors: | Owen, C.D, Wu, H, Crost, E, Colvile, A, Juge, N, Walsh, M.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fucosidases from the human gut symbiont Ruminococcus gnavus.
Cell.Mol.Life Sci., 78, 2021
|
|
1EUT
 
 | | SIALIDASE, LARGE 68KD FORM, COMPLEXED WITH GALACTOSE | | Descriptor: | SIALIDASE, SODIUM ION | | Authors: | Gaskell, A, Crennell, S.J, Taylor, G.L. | | Deposit date: | 1996-06-21 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll.
Structure, 3, 1995
|
|
3J2S
 
 | |
1GOH
 
 | |
1GOG
 
 | |
1GOF
 
 | | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | | Descriptor: | ACETIC ACID, COPPER (II) ION, GALACTOSE OXIDASE, ... | | Authors: | Ito, N, Phillips, S.E.V, Knowles, P.F. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase.
Nature, 350, 1991
|
|
3MO4
 
 | | The crystal structure of an alpha-(1-3,4)-fucosidase from Bifidobacterium longum subsp. infantis ATCC 15697 | | Descriptor: | Alpha-1,3/4-fucosidase, FORMIC ACID, TYROSINE | | Authors: | Tan, K, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-12 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides.
Appl.Environ.Microbiol., 78, 2012
|
|
