7RNX
 
 | |
7N9W
 
 | |
7N8P
 
 | |
7JM7
 
 | | Structure of human CLC-7/OSTM1 complex | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3S,4S,5R,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dinonanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schrecker, M, Hite, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1.
Elife, 9, 2020
|
|
7JM6
 
 | | Structure of chicken CLC-7 | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1S,2R,3S,4S,5R,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dinonanoate, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schrecker, M, Hite, R. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of the lysosomal chloride-proton exchanger CLC-7 in complex with OSTM1.
Elife, 9, 2020
|
|
7CVT
 
 | | Crystal structure of the C85A/L194A/H234C mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
7CVS
 
 | | Crystal structure of the C85A/L194A mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
7CQ7
 
 | | Structure of the human CLCN7-OSTM1 complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ADP
To Be Published
|
|
7CQ6
 
 | | Structure of the human CLCN7-OSTM1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, H(+)/Cl(-) exchange transporter 7, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex
To Be Published
|
|
7CQ5
 
 | | Structure of the human CLCN7-OSTM1 complex with ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ATP
To Be Published
|
|
7BXU
 
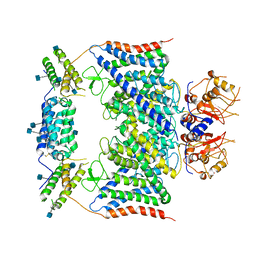 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
6V2J
 
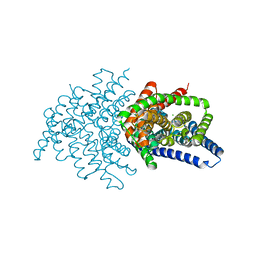 | | Crystal structure of ClC-ec1 triple mutant (E113Q, E148Q, E203Q) | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA | | Authors: | Maduke, M, Mathews, I.I, Chavan, T.S. | | Deposit date: | 2019-11-24 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A CLC-ec1 mutant reveals global conformational change and suggests a unifying mechanism for the CLC Cl - /H + transport cycle.
Elife, 9, 2020
|
|
6QVU
 
 | |
6QVD
 
 | |
6QVC
 
 | |
6QVB
 
 | |
6QV6
 
 | |
6LSC
 
 | |
6K5F
 
 | | Crystal structure of the CLC-ec1 deltaNC in presence of 200 mM NaBr | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Park, K, Lim, H.H. | | Deposit date: | 2019-05-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K5D
 
 | |
6COZ
 
 | |
6COY
 
 | |
6ADC
 
 | |
6ADB
 
 | | Crystal structure of the E148N mutant CLC-ec1 in 20mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ADA
 
 | | Crystal structure of the E148D mutant CLC-ec1 in 200mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
