1BOI
 
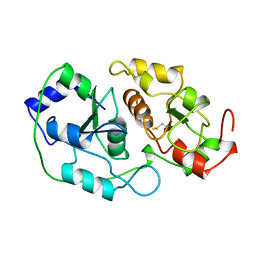 | | N-TERMINALLY TRUNCATED RHODANESE | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1C25
 
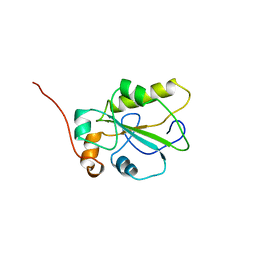 | | HUMAN CDC25A CATALYTIC DOMAIN | | Descriptor: | CDC25A | | Authors: | Fauman, E.B, Cogswell, J.P, Lovejoy, B, Rocque, W.J, Holmes, W, Montana, V.G, Piwnica-Worms, H, Rink, M.J, Saper, M.A. | | Deposit date: | 1998-04-17 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A.
Cell(Cambridge,Mass.), 93, 1998
|
|
5WQJ
 
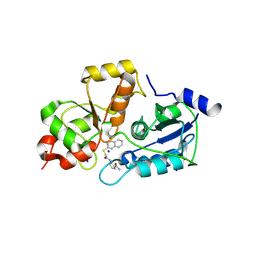 | |
5WQK
 
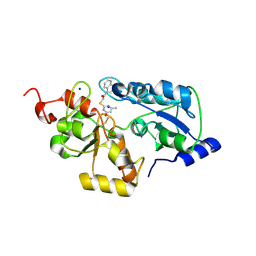 | |
3TG3
 
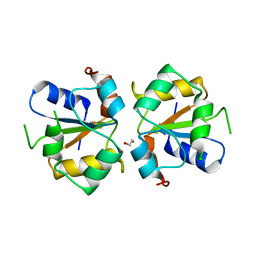 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
2MOI
 
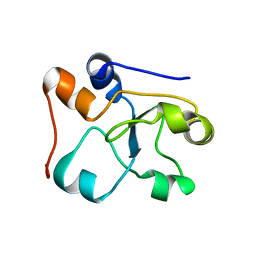 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2MOL
 
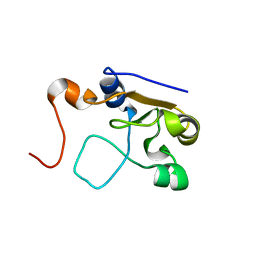 | | 3D NMR structure of the cytoplasmic rhodanese domain of the full-length inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-27 | | Release date: | 2014-06-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
2VSW
 
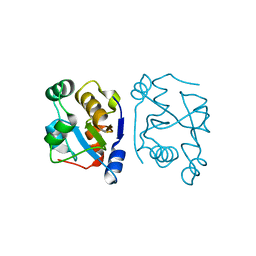 | | The structure of the rhodanese domain of the human dual specificity phosphatase 16 | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 16 | | Authors: | Murray, J.W, Barr, A, Pike, A.C.W, Elkins, J, Phillips, C, Wang, J, Savitsky, P, Roos, A, Bishop, S, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Burgess-Brown, N, Pantic, N, Bray, J, von Delft, F, Gileadi, O, Knapp, S. | | Deposit date: | 2008-04-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Rhodanese Domain of the Human Dual Specifity Phosphatase 16
To be Published
|
|
2ORA
 
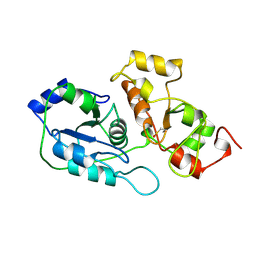 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
2UZQ
 
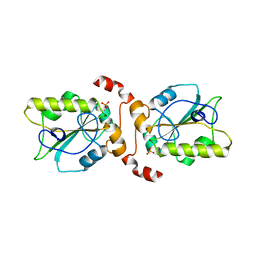 | | Protein Phosphatase, New Crystal Form | | Descriptor: | M-PHASE INDUCER PHOSPHATASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U. | | Deposit date: | 2007-05-01 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | New Crystal Form of Protein Phosphatase Cdc25B Triggered by Guanidinium Chloride as an Additive
To be Published
|
|
2WLR
 
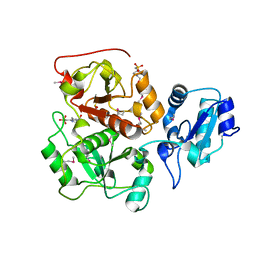 | |
2WLX
 
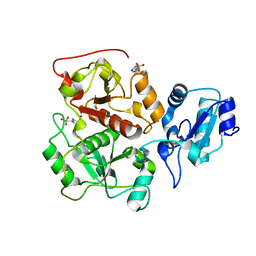 | |
2OUC
 
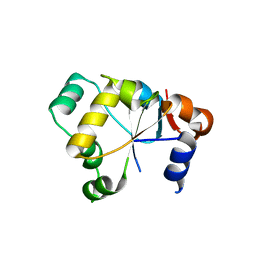 | |
5HBO
 
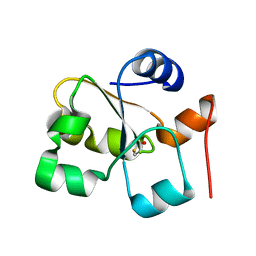 | | Native rhodanese domain of YgaP prepared without DDT is both S-nitrosylated and S-sulfhydrated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HPA
 
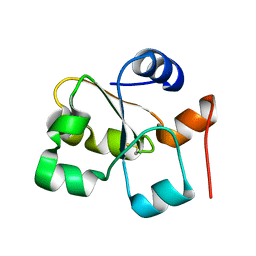 | |
5HBL
 
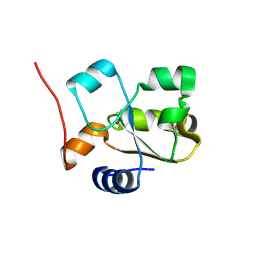 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBP
 
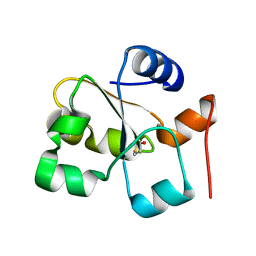 | | The crystal of rhodanese domain of YgaP treated with SNOC | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-01 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5HBQ
 
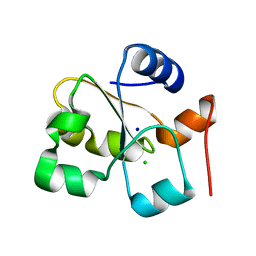 | | C63D mutant of the rhodanese domain of YgaP | | Descriptor: | CHLORIDE ION, Inner membrane protein YgaP, SODIUM ION | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2016-01-02 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
1DP2
 
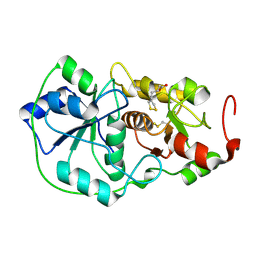 | |
1E0C
 
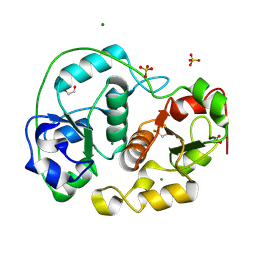 | | SULFURTRANSFERASE FROM AZOTOBACTER VINELANDII | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bordo, D, Deriu, D, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-08 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of a Sulfurtransferase from Azotobacter Vinelandii Highlights the Evolutionary Relationship between the Rhodanese and Phosphatase Enzyme Families
J.Mol.Biol., 298, 2000
|
|
3FOJ
 
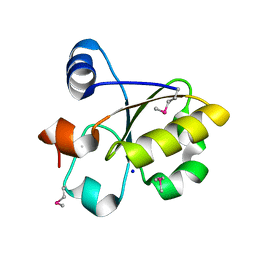 | | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A. | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Seetharaman, J, Abashidze, M, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-30 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of SSP1007 From Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Target SyR101A.
To be Published
|
|
3F4A
 
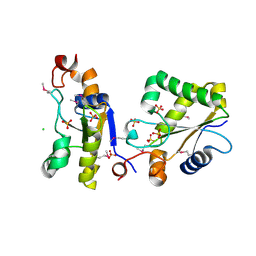 | | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family | | Descriptor: | AMMONIUM ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Osipiuk, J, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ygr203w, a yeast protein tyrosine phosphatase of the Rhodanese family
To be Published
|
|
3FLH
 
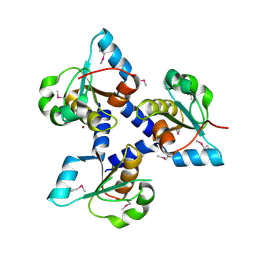 | | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B | | Descriptor: | BROMIDE ION, uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B
To be Published
|
|
3D1P
 
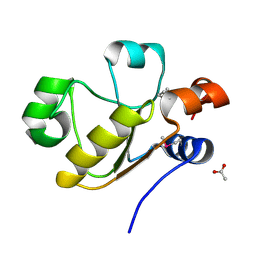 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | Authors: | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
1CWR
 
 | |
